bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-8_CDS_annotation_glimmer3.pl_2_6
Length=290
Score E
Sequences producing significant alignments: (Bits) Value
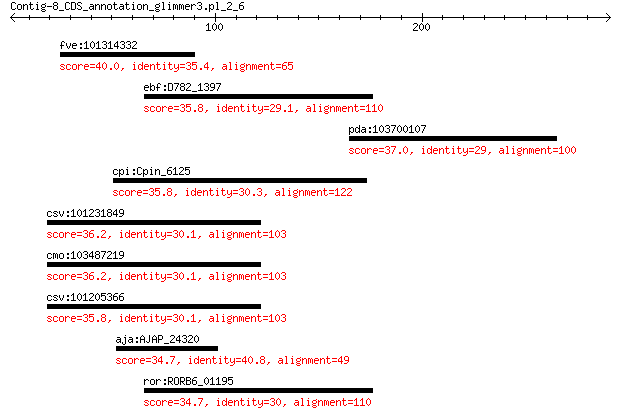
fve:101314332 capsid protein VP1-like 40.0 0.40
ebf:D782_1397 putative acyltransferase 35.8 3.9
pda:103700107 uncharacterized LOC103700107 37.0 4.2
cpi:Cpin_6125 uracil-DNA glycosylase 35.8 5.5
csv:101231849 floral homeotic protein AGAMOUS-like 36.2 6.2
cmo:103487219 floral homeotic protein AGAMOUS-like 36.2 6.4
csv:101205366 floral homeotic protein AGAMOUS-like 35.8 7.3
aja:AJAP_24320 Hypothetical protein 34.7 7.8
ror:RORB6_01195 elaA protein 34.7 9.8
> fve:101314332 capsid protein VP1-like
Length=421
Score = 40.0 bits (92), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 29/65 (45%), Gaps = 0/65 (0%)
Query 25 VTSGISINDLRNVNAYQKFLELNMRKGYSYRDIIEGRFNVKVRYDELLMPEFFGGFSRDI 84
+ +IN LR QK LE + R G Y +II F V L PE+ GG S I
Sbjct 302 AATAATINQLRQSFQIQKLLERDARGGTRYTEIIRSHFGVASPDARLQRPEYLGGGSTPI 361
Query 85 EMHSI 89
+ I
Sbjct 362 NIAPI 366
> ebf:D782_1397 putative acyltransferase
Length=153
Score = 35.8 bits (81), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 32/130 (25%), Positives = 53/130 (41%), Gaps = 24/130 (18%)
Query 66 VRYDELLMPEFFGGFSRDIEMHSISQT-VDQDLDGSQTYAKALGSQSGIAGVRGDS---- 120
+ + +L +P+ + E+ + Q V QD+DG +G + G +GD+
Sbjct 7 LHHSDLTVPQLYALLQLRCEVFVVEQACVYQDIDGDDL----VGENRHLLGWQGDTLVAY 62
Query 121 GRALECFCDEESIVMGILIVTP---------------LPVYTQLLPKHFTYRGLLDHYQP 165
R L+ D + +G +IV+P L Q P+H Y G H Q
Sbjct 63 ARLLKSEDDFSPVSIGRVIVSPAVRGEKLGYQLMERALASCAQHWPEHALYLGAQAHLQN 122
Query 166 EFNHIGFQPI 175
+ H GF P+
Sbjct 123 FYRHFGFIPV 132
> pda:103700107 uncharacterized LOC103700107
Length=332
Score = 37.0 bits (84), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 29/102 (28%), Positives = 50/102 (49%), Gaps = 7/102 (7%)
Query 165 PEFNHIGFQPILYKEVCPLQ-AYSDGPDTLSDVFGYNRPWYEYVQKYDQAHGLFRTNLSN 223
P+F + P + + CPL + S GP S F + + W Y Q +D +RT +
Sbjct 152 PQFTWVSHLPRIASDYCPLLISTSSGPCHHSP-FRFEKVWLSYPQSWDIVRDAWRTPVRG 210
Query 224 FLMHRVFNQKPQLAQSFL-VIDPAQVTDVFAVTKADDGTELA 264
MH++ ++K +LA+ L + V D+F + +G E+A
Sbjct 211 DAMHQI-SRKLELARRHLRRWNREVVGDIF---QRVEGVEIA 248
> cpi:Cpin_6125 uracil-DNA glycosylase
Length=171
Score = 35.8 bits (81), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 37/125 (30%), Positives = 50/125 (40%), Gaps = 24/125 (19%)
Query 51 GYSYRDIIEGRFNVKVRYDELLMPEF---FGGFSRDIEMHSISQTVDQDLDGSQTYAKAL 107
GY +E VR DEL EF F F R IE + TY L
Sbjct 66 GYGLTTAVE---RATVRADELSKAEFDQSFETFKRKIEQYK------------PTYLAFL 110
Query 108 GSQSGIAGVRGDSGRALECFCDEESIVMGILIVTPLPVYTQLLPKHFTYRGLLDHYQPEF 167
G + +A + ++ EE + I+ V P T L + FT L+DH+Q
Sbjct 111 GKAAYMAF---SEKKNVQWGLQEEPLCNAIVWVLP---NTSGLNRGFTLSMLIDHFQALH 164
Query 168 NHIGF 172
NHIG+
Sbjct 165 NHIGY 169
> csv:101231849 floral homeotic protein AGAMOUS-like
Length=262
Score = 36.2 bits (82), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 31/106 (29%), Positives = 50/106 (47%), Gaps = 20/106 (19%)
Query 19 RNLIDVVTSGISINDLRNVNAYQKFLELNMRKGYSYRDIIEGRFNVKVRYDELLMPEFFG 78
R+L+ S +S+ DL K LE+ + KG S ++ R +ELL E
Sbjct 141 RHLLGESISSLSVKDL-------KSLEVKLEKGISR---------IRSRKNELLFSEIEY 184
Query 79 GFSRDIEMHSISQTVDQDL---DGSQTYAKALGSQSGIAGVRGDSG 121
R+IE+H+ +Q + + + SQ A + +GIA RG+ G
Sbjct 185 MQKREIELHTNNQLIRAKIAETERSQQNTNA-SNNNGIATRRGEEG 229
> cmo:103487219 floral homeotic protein AGAMOUS-like
Length=265
Score = 36.2 bits (82), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 31/106 (29%), Positives = 50/106 (47%), Gaps = 19/106 (18%)
Query 19 RNLIDVVTSGISINDLRNVNAYQKFLELNMRKGYSYRDIIEGRFNVKVRYDELLMPEFFG 78
R+L+ S +S+ DL K LE+ + KG S ++ R +ELL E
Sbjct 142 RHLLGESISSLSVKDL-------KSLEVKLEKGLS---------RIRSRKNELLFSEIEY 185
Query 79 GFSRDIEMHSISQTVDQDL-DGSQTYAKALGSQSGIA--GVRGDSG 121
R+IE+H+ +Q + + + ++ S +GIA G RGD G
Sbjct 186 MQKREIELHTNNQLIRAKIAETERSQQNRNASNNGIAATGGRGDEG 231
> csv:101205366 floral homeotic protein AGAMOUS-like
Length=254
Score = 35.8 bits (81), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 31/106 (29%), Positives = 50/106 (47%), Gaps = 20/106 (19%)
Query 19 RNLIDVVTSGISINDLRNVNAYQKFLELNMRKGYSYRDIIEGRFNVKVRYDELLMPEFFG 78
R+L+ S +S+ DL K LE+ + KG S ++ R +ELL E
Sbjct 133 RHLLGESISSLSVKDL-------KSLEVKLEKGISR---------IRSRKNELLFSEIEY 176
Query 79 GFSRDIEMHSISQTVDQDL---DGSQTYAKALGSQSGIAGVRGDSG 121
R+IE+H+ +Q + + + SQ A + +GIA RG+ G
Sbjct 177 MQKREIELHTNNQLIRAKIAETERSQQNTNA-SNNNGIATRRGEEG 221
> aja:AJAP_24320 Hypothetical protein
Length=132
Score = 34.7 bits (78), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 20/49 (41%), Positives = 28/49 (57%), Gaps = 2/49 (4%)
Query 52 YSYRDIIEGRFNVKVRYDELLMPEFFGGFSRDIEMHSISQTVDQDLDGS 100
Y+ R I GR+NVK+ Y ++ FFG S I++ TV QD +GS
Sbjct 42 YAGRHIGNGRYNVKITYVDV--ARFFGSQSGTIDITMADGTVLQDFNGS 88
> ror:RORB6_01195 elaA protein
Length=153
Score = 34.7 bits (78), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 33/130 (25%), Positives = 51/130 (39%), Gaps = 24/130 (18%)
Query 66 VRYDELLMPEFFGGFSRDIEMHSISQTVD-QDLDGSQTYAKALGSQSGIAGVRGDS---- 120
+ + EL +P+ + + + QT QD+DG +G + G RGD
Sbjct 7 LHHAELTVPQLYALLQLRCAVFVVEQTCPYQDVDGDDL----IGENRHLLGWRGDELVAY 62
Query 121 GRALECFCDEESIVMGILIVTP---------------LPVYTQLLPKHFTYRGLLDHYQP 165
R L+ D E +V+G +IV+P L + P Y G H Q
Sbjct 63 ARILKSDDDIEPVVIGRVIVSPAVRGEKLGYQLMERALASCRRHWPHKAIYLGAQAHLQR 122
Query 166 EFNHIGFQPI 175
+ GFQP+
Sbjct 123 FYASFGFQPV 132
Lambda K H a alpha
0.322 0.141 0.422 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 479831790136