bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-8_CDS_annotation_glimmer3.pl_2_4
Length=368
Score E
Sequences producing significant alignments: (Bits) Value
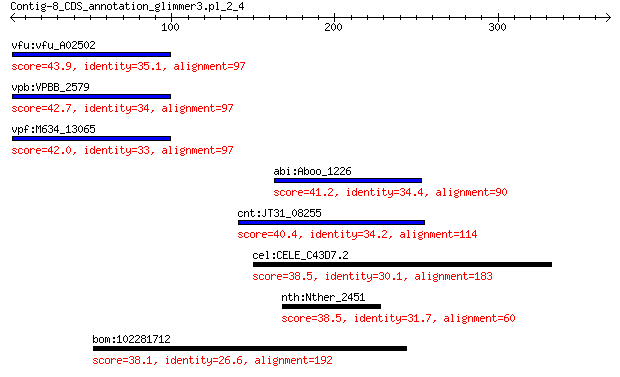
vfu:vfu_A02502 bacteriophage protein 43.9 0.038
vpb:VPBB_2579 putative bacteriophage protein 42.7 0.080
vpf:M634_13065 hypothetical protein 42.0 0.18
abi:Aboo_1226 CUB protein 41.2 0.39
cnt:JT31_08255 LysR family transcriptional regulator 40.4 0.41
cel:CELE_C43D7.2 fbxb-65; Protein FBXB-65 38.5 1.8
nth:Nther_2451 hypothetical protein 38.5 1.8
bom:102281712 CEP164; centrosomal protein 164kDa 38.1 3.3
> vfu:vfu_A02502 bacteriophage protein
Length=395
Score = 43.9 bits (102), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 34/105 (32%), Positives = 50/105 (48%), Gaps = 18/105 (17%)
Query 2 YGLIGAAIGAISQNFNTSESIAANKQEQQSNRDYNLNLARLQNQWNREQWEREAQYNSPA 61
Y ++ AI I+ N +E + +++SN + L + R QW A+++ A
Sbjct 175 YHILPTAIPGIAAATNPAEWLLEAGADKESNDELRLRI--------RNQWSAVARWHIDA 226
Query 62 AYRARLAS-AGMNSDLAYSNVNGVAPASPG-------MTSGEPSS 98
AYRA L S AG+N D Y N AP PG + +GEPS
Sbjct 227 AYRALLTSRAGINDDNVYFEHN--APRGPGTANALILLDTGEPSP 269
> vpb:VPBB_2579 putative bacteriophage protein
Length=395
Score = 42.7 bits (99), Expect = 0.080, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 51/105 (49%), Gaps = 18/105 (17%)
Query 2 YGLIGAAIGAISQNFNTSESIAANKQEQQSNRDYNLNLARLQNQWNREQWEREAQYNSPA 61
Y ++ AI I+ N +E + +++SN + L + R QW A+++ A
Sbjct 175 YHILPTAIPGIAAATNPAEWLTQAGADKESNDELRLRV--------RNQWSAVARWHIDA 226
Query 62 AYRARLAS-AGMNSDLAYSNVNGVAPASPG-------MTSGEPSS 98
AYR+ L S AG+N D Y N AP PG + +GEPS+
Sbjct 227 AYRSLLTSRAGINDDNVYFQHN--APRGPGTANALILLDTGEPSA 269
> vpf:M634_13065 hypothetical protein
Length=395
Score = 42.0 bits (97), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 32/105 (30%), Positives = 50/105 (48%), Gaps = 18/105 (17%)
Query 2 YGLIGAAIGAISQNFNTSESIAANKQEQQSNRDYNLNLARLQNQWNREQWEREAQYNSPA 61
+ ++ AI I+ N +E + +++SN + L + R QW A+++ A
Sbjct 175 FHILPTAIPGIAAATNPAEWLTEAGADKESNDELRLRI--------RNQWSAVAKWHIDA 226
Query 62 AYRARLAS-AGMNSDLAYSNVNGVAPASPG-------MTSGEPSS 98
AYR+ L S AG+N D Y N AP PG + +GEPS
Sbjct 227 AYRSLLTSRAGINDDNVYFEHN--APRGPGTANSYILLDTGEPSP 269
> abi:Aboo_1226 CUB protein
Length=1621
Score = 41.2 bits (95), Expect = 0.39, Method: Composition-based stats.
Identities = 31/97 (32%), Positives = 46/97 (47%), Gaps = 21/97 (22%)
Query 163 NTLEIQLKEGVIKLNDSVKQLN-------EQNKKNLQQLLENLKLESNNLAEQWQVIRET 215
NTL+ LKE V LN ++++LN EQN NLQ + +LK ++N+L Q I
Sbjct 1481 NTLQNDLKENVTALNKAIEELNTTLVNKIEQNINNLQSQINDLKNQANDLQSNIQQINTN 1540
Query 216 WSNLRVDRALKMIDLKFREKQNVAILKKISSETNLNY 252
S++ + +N K ISS N+ Y
Sbjct 1541 ISDI--------------QSKNNEQDKAISSSNNIGY 1563
> cnt:JT31_08255 LysR family transcriptional regulator
Length=300
Score = 40.4 bits (93), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 39/127 (31%), Positives = 65/127 (51%), Gaps = 18/127 (14%)
Query 141 EAGKTSEEAEGVRIDNLTRGASNTLE-IQLKEGVIKLNDSVKQLN-----EQNKKNLQQL 194
E+G S AE + + N S T++ +++K GV LN + +QL+ E+ +N+QQ+
Sbjct 16 ESGSFSRAAEQLGLAN--SAVSRTVKKLEMKLGVSLLNRTTRQLSLTEEGERYFRNVQQI 73
Query 195 LENLKLESNNLAEQWQVIRETWSNLRVDRALK-MIDL------KFREKQNVAILKKISSE 247
L+ + N + E Q+ R LR+D A M+ L FRE+ L +SSE
Sbjct 74 LQAMAAAENEVMESKQMPR---GLLRIDAATPVMLHLLMPLIKPFRERYPEMTLSLVSSE 130
Query 248 TNLNYVQ 254
T +N ++
Sbjct 131 TFINLIE 137
> cel:CELE_C43D7.2 fbxb-65; Protein FBXB-65
Length=307
Score = 38.5 bits (88), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 55/203 (27%), Positives = 93/203 (46%), Gaps = 46/203 (23%)
Query 150 EGVRIDNLTRGAS----NTLEIQLKEGVIKLNDSVKQLNEQNKKNLQQLLENLKLESNNL 205
E V ID L R S N LE++L+ KL D +K + NK LE L ++S+N
Sbjct 59 EFVSID-LRRSKSLIKWNPLEVELR----KLIDHIKSVFHTNK------LEVLIIQSDN- 106
Query 206 AEQWQVIRETWSNLRVDRA-LKMIDLKFREKQNVAILKKIS--SETNLNYVQASSM---- 258
W+ + E L R L++ + QN + K I+ S+ +L+ + +SS
Sbjct 107 -HDWKSVHEALDGLDCSRTGLEV------QSQNRYLQKIINKFSKRDLDLIVSSSTWYHP 159
Query 259 ---TKRLMLDMALGKTQM------NLMTQQAITETQKRVNMRTEDWYNRGRIRSVYLQNG 309
+ R L + G T + L+ QQ +T + +N+ + W I+ YL
Sbjct 160 ILSSNRKYLTLTAGFTDLLCVNCKYLIVQQPVT--ARDLNLMLKHW-----IKGAYLSVH 212
Query 310 QLSFDLSQSIKWDDTMKSIDWCE 332
QL ++ I++DD K++++ E
Sbjct 213 QLEVAVNSRIRFDDVFKNVEYTE 235
> nth:Nther_2451 hypothetical protein
Length=321
Score = 38.5 bits (88), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 19/64 (30%), Positives = 41/64 (64%), Gaps = 5/64 (8%)
Query 168 QLKEGVIKLNDSVKQLNEQNKKNLQQLLENLKLESNNLAEQ----WQVIRETWSNLRVDR 223
Q+K+G++++ ++V++ ++ + +Q+ +N+K E N AEQ + IRE+ + +R D
Sbjct 100 QVKDGIVQIGETVQETTQEVTETIQETRDNVKTEVNKTAEQVEHSYDEIRESATEIR-DV 158
Query 224 ALKM 227
A +M
Sbjct 159 AGEM 162
> bom:102281712 CEP164; centrosomal protein 164kDa
Length=1388
Score = 38.1 bits (87), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 51/209 (24%), Positives = 93/209 (44%), Gaps = 26/209 (12%)
Query 52 EREAQYNSPAAY--------RARLASAGMNSDLAYSNVNGVAPASPGMTSGEPSSPVDYS 103
+REA + PAA+ +A G V+ P SP + S EP++P +
Sbjct 520 QREALPSPPAAHERGQEWRPQAEGPGPGQEEPKEKVAVSPTPPGSPEVRSAEPAAPREQL 579
Query 104 AIAGKQTIGSAVSQALANEQARANIALTQAQKNKTDEEAGKTSEEAEGVRIDNLTRGASN 163
+ A + AV++ L EQ R + ++ +K + E EEAE +++ A +
Sbjct 580 SEAALEATDEAVTRELEQEQTR--LLESKQEKMQRLREKLCREEEAEALQLHQQKEKALS 637
Query 164 TLEIQLKEGVIKLNDSVKQLNEQNKKNLQQLLENLKLESNNLAEQ--------WQVIRET 215
+L+ QL+ + Q+ EQ ++ L +L ++ S A+Q Q +RE
Sbjct 638 SLKEQLQRAT---EEEETQMREQERQRLSRLRAQVQSSSEADADQIRAEQEASLQRLREE 694
Query 216 WSNL-RVDRALKMIDLKFREKQNVAILKK 243
+L + +RA L+ R +Q + L++
Sbjct 695 LESLQKAERA----SLEERSRQTLEQLRE 719
Lambda K H a alpha
0.311 0.124 0.347 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 706893140446