bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-7_CDS_annotation_glimmer3.pl_2_3
Length=71
Score E
Sequences producing significant alignments: (Bits) Value
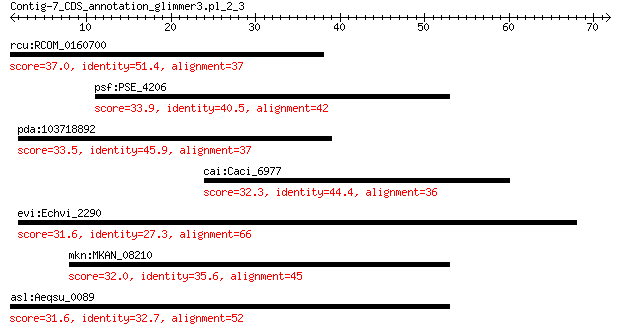
rcu:RCOM_0160700 pentatricopeptide repeat-containing protein, ... 37.0 0.22
psf:PSE_4206 glutamine synthetase 33.9 2.3
pda:103718892 pentatricopeptide repeat-containing protein At4g... 33.5 3.1
cai:Caci_6977 hypothetical protein 32.3 8.3
evi:Echvi_2290 cAMP-binding protein 31.6 8.6
mkn:MKAN_08210 glutamine amidotransferase 32.0 8.8
asl:Aeqsu_0089 small-conductance mechanosensitive channel 31.6 9.8
> rcu:RCOM_0160700 pentatricopeptide repeat-containing protein,
putative
Length=726
Score = 37.0 bits (84), Expect = 0.22, Method: Composition-based stats.
Identities = 19/44 (43%), Positives = 29/44 (66%), Gaps = 7/44 (16%)
Query 1 MKENFYFVL-----YLGTSVLNVY--AGSQSNARAYFRRWIEKN 37
+ NF+++L +LGTS+LN+Y GS S+AR YF R + K+
Sbjct 455 LTRNFFYILEGDNIHLGTSILNMYIRCGSISSAREYFNRMVAKD 498
> psf:PSE_4206 glutamine synthetase
Length=468
Score = 33.9 bits (76), Expect = 2.3, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 24/42 (57%), Gaps = 6/42 (14%)
Query 11 LGTSVLNVYAGSQSNARAYFRRWIEKNLPVYYHESDFWFLRY 52
LG ++ + YA S+ N FR WIEK++ + F FLRY
Sbjct 429 LGVALADQYARSRENELKAFRAWIEKDI------TSFEFLRY 464
> pda:103718892 pentatricopeptide repeat-containing protein At4g30700-like
Length=649
Score = 33.5 bits (75), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 17/39 (44%), Positives = 25/39 (64%), Gaps = 2/39 (5%)
Query 2 KENFYFVLYLGTSVLNVYA--GSQSNARAYFRRWIEKNL 38
+ NF F + LGT+++++YA GS AR F R EKN+
Sbjct 305 RRNFSFDVILGTAMIDMYARCGSMDAAREIFDRMKEKNV 343
> cai:Caci_6977 hypothetical protein
Length=897
Score = 32.3 bits (72), Expect = 8.3, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 22/43 (51%), Gaps = 7/43 (16%)
Query 24 SNARAYFRRWIEK-------NLPVYYHESDFWFLRYECFSDFL 59
S +R+Y +W+ K NL + Y S W L+Y FSD L
Sbjct 608 STSRSYISQWVSKGEDPAGTNLDLAYGSSTTWSLKYNGFSDQL 650
> evi:Echvi_2290 cAMP-binding protein
Length=218
Score = 31.6 bits (70), Expect = 8.6, Method: Composition-based stats.
Identities = 18/71 (25%), Positives = 35/71 (49%), Gaps = 7/71 (10%)
Query 2 KENFYFVLYLGTSVLNVYAGSQSNARAYFRRWIEKN-----LPVYYHESDFWFLRYECFS 56
KE F + LY G + ++ + + +E++ +PV YHE W+ +Y+ +
Sbjct 65 KELFLYYLYPGETCALSLTCCEAGKASEIKAIVEEDTKVLLVPVQYHEQ--WYEQYKQWK 122
Query 57 DFLASRYPVKF 67
DF+ + Y +F
Sbjct 123 DFVTATYQKRF 133
> mkn:MKAN_08210 glutamine amidotransferase
Length=284
Score = 32.0 bits (71), Expect = 8.8, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 23/51 (45%), Gaps = 6/51 (12%)
Query 8 VLYLGTSVLNVYAGSQSNARAYFRRWIEKNLPVY------YHESDFWFLRY 52
V L T+ + + G QS RW+ N+P+Y +D W LRY
Sbjct 137 VFALITATIRAHGGDQSAGLVDALRWLAANVPIYAVNVLLSTATDMWALRY 187
> asl:Aeqsu_0089 small-conductance mechanosensitive channel
Length=373
Score = 31.6 bits (70), Expect = 9.8, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 29/53 (55%), Gaps = 3/53 (6%)
Query 1 MKENFYFVLYLG-TSVLNVYAGSQSNARAYFRRWIEKNLPVYYHESDFWFLRY 52
+K NF +LYLG +V+ + + +N +FR I++ + + F FLRY
Sbjct 84 LKHNFNIILYLGIVAVVTIVGAASTN--MWFRMSIKRKIENLEDSTSFKFLRY 134
Lambda K H a alpha
0.329 0.142 0.465 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 126269703504