bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-6_CDS_annotation_glimmer3.pl_2_3
Length=382
Score E
Sequences producing significant alignments: (Bits) Value
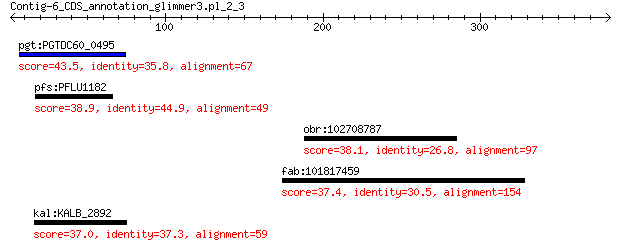
pgt:PGTDC60_0495 hypothetical protein 43.5 0.009
pfs:PFLU1182 hypothetical protein 38.9 1.5
obr:102708787 125 kDa kinesin-related protein-like 38.1 2.9
fab:101817459 LRRCC1; leucine rich repeat and coiled-coil cent... 37.4 6.3
kal:KALB_2892 hypothetical protein 37.0 7.9
> pgt:PGTDC60_0495 hypothetical protein
Length=103
Score = 43.5 bits (101), Expect = 0.009, Method: Composition-based stats.
Identities = 24/69 (35%), Positives = 39/69 (57%), Gaps = 5/69 (7%)
Query 7 GAAAGAAAGSVVPGIGTAIGAIGGAAISAIGNWFGNRSNRKASREA--FERESKFAREER 64
GAA GAA GSV+PG+GT G I GAA+ ++ G ++N A F ++++F + +
Sbjct 37 GAAIGAAIGSVIPGLGTITGGIAGAAVESL---IGEKANENIDNFADNFFQDTEFEFQCK 93
Query 65 LAQQQWIEQ 73
+W +
Sbjct 94 SCSHKWTRK 102
> pfs:PFLU1182 hypothetical protein
Length=383
Score = 38.9 bits (89), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 22/54 (41%), Positives = 31/54 (57%), Gaps = 5/54 (9%)
Query 17 VVPGIGTAI-GAIGGAAISAIGNWFGNR----SNRKASREAFERESKFAREERL 65
V+PG+GTA+ GAIGG S G W G++ S+R S A +E AR + +
Sbjct 275 VLPGVGTAVGGAIGGLLGSEAGAWLGDKLFGSSDRLPSPNAVSKELNHARPDNV 328
> obr:102708787 125 kDa kinesin-related protein-like
Length=623
Score = 38.1 bits (87), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 26/104 (25%), Positives = 54/104 (52%), Gaps = 16/104 (15%)
Query 188 IYVNHELGQLNHAEAEVAAKKLQEIDVAMSEARERISTMK----AQQSQIDENMVQLKFD 243
IY+ HE L AE + K++ +++++ + R+ + K A+Q + D
Sbjct 437 IYIPHERFALEEAEKKTMKDKIESLELSIEDHRKEVDKFKRLYLAEQEH--------RLD 488
Query 244 RYLRSKEFEL---LCKRTYQDMKESNSRINLNAAEVQDMMATQL 284
R+KE ++ CK+ + D++E++SR N++ E +D + + L
Sbjct 489 LESRNKELKMNIESCKKEFLDLEEAHSRANISLKE-KDFIISNL 531
> fab:101817459 LRRCC1; leucine rich repeat and coiled-coil centrosomal
protein 1
Length=943
Score = 37.4 bits (85), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 47/170 (28%), Positives = 81/170 (48%), Gaps = 17/170 (10%)
Query 174 KARTESDIE-LNNSTIYVNHELGQLNHAEAEVAAKKLQEIDVAMSEAR-ERISTMKAQQS 231
KAR ++DI+ L T + +EL Q + EAE K LQ ++ +S+ +R+ A+
Sbjct 482 KARLQADIQQLKGETERLTNELKQAKNKEAEYQ-KTLQALEETLSKMEMQRLQQRAAEMK 540
Query 232 QIDE---------NMVQLKFDRYLRSKE-----FELLCKRTYQDMKESNSRINLNAAEVQ 277
Q+ E VQL + KE ELL R + KE SR+ LN E Q
Sbjct 541 QVQEAELKSSANAREVQLLRISVRQQKEKVKHLHELLVLREQEQRKELGSRVALNGPEFQ 600
Query 278 DMMATQLARVMNLNASTYMQKKQGILASEQTMTELYKQTGIDISNQHAKF 327
D ++ ++A+ +A + ++ I S+Q EL ++ + ++ + +F
Sbjct 601 DALSKEVAKEEQRHAHHVRELQEKINISDQKYKELEQEFHLALTIEAKRF 650
> kal:KALB_2892 hypothetical protein
Length=632
Score = 37.0 bits (84), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 34/59 (58%), Gaps = 5/59 (8%)
Query 16 SVVPGIGTAIGAIGGAAISAIGNWFGNRSNRKASREAFERESKFAREERLAQQQWIEQM 74
S++PGIG AIG + G SA+G FG R+ + S EA + + R+ LA + +E +
Sbjct 490 SLLPGIGAAIGGVLG---SALGTLFGARNGYRTSMEASREQQR--RQRALALRHELEPL 543
Lambda K H a alpha
0.311 0.124 0.341 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 750490074218