bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-38_CDS_annotation_glimmer3.pl_2_1
Length=90
Score E
Sequences producing significant alignments: (Bits) Value
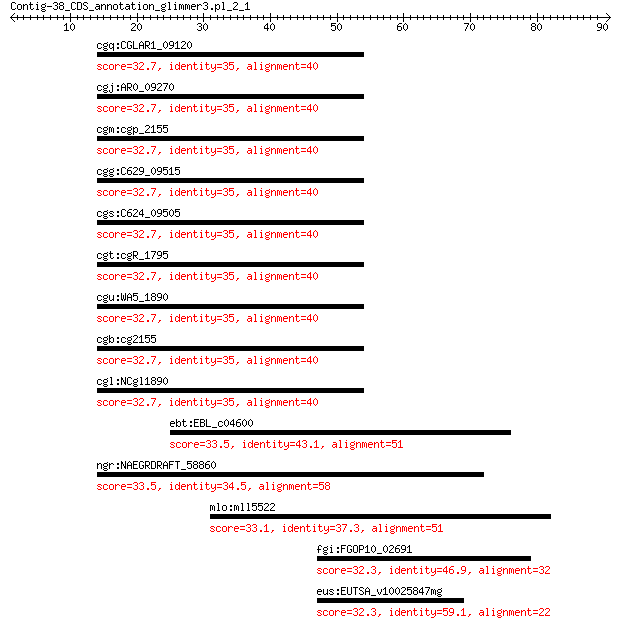
cgq:CGLAR1_09120 hypothetical protein 32.7 2.5
cgj:AR0_09270 hypothetical protein 32.7 2.5
cgm:cgp_2155 hypothetical protein 32.7 2.5
cgg:C629_09515 hypothetical protein 32.7 2.5
cgs:C624_09505 hypothetical protein 32.7 2.5
cgt:cgR_1795 hypothetical protein 32.7 2.5
cgu:WA5_1890 hypothetical protein 32.7 2.5
cgb:cg2155 hypothetical protein 32.7 2.5
cgl:NCgl1890 Cgl1965; hypothetical protein 32.7 2.5
ebt:EBL_c04600 exuR; negative regulator of exu regulon 33.5 3.8
ngr:NAEGRDRAFT_58860 peptidase C2, calpain 33.5 3.8
mlo:mll5522 rubredoxin reductase 33.1 5.8
fgi:FGOP10_02691 uracil-DNA glycosylase superfamily protein 32.3 9.3
eus:EUTSA_v10025847mg hypothetical protein 32.3 9.3
> cgq:CGLAR1_09120 hypothetical protein
Length=95
Score = 32.7 bits (73), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 21/40 (53%), Gaps = 0/40 (0%)
Query 14 IEAMSNLIQAEAAKNSGLAAVENAATNRAQQAWRELTTQY 53
+E SNL+ AE N+ VEN NR ++W +T +
Sbjct 56 LEEGSNLVDAETLMNNDPFHVENVLDNRVIRSWNPVTKDF 95
> cgj:AR0_09270 hypothetical protein
Length=95
Score = 32.7 bits (73), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 21/40 (53%), Gaps = 0/40 (0%)
Query 14 IEAMSNLIQAEAAKNSGLAAVENAATNRAQQAWRELTTQY 53
+E SNL+ AE N+ VEN NR ++W +T +
Sbjct 56 LEEGSNLVDAETLMNNDPFHVENVLDNRVIRSWNPVTKDF 95
> cgm:cgp_2155 hypothetical protein
Length=95
Score = 32.7 bits (73), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 21/40 (53%), Gaps = 0/40 (0%)
Query 14 IEAMSNLIQAEAAKNSGLAAVENAATNRAQQAWRELTTQY 53
+E SNL+ AE N+ VEN NR ++W +T +
Sbjct 56 LEEGSNLVDAETLMNNDPFHVENVLDNRVIRSWNPVTKDF 95
> cgg:C629_09515 hypothetical protein
Length=95
Score = 32.7 bits (73), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 21/40 (53%), Gaps = 0/40 (0%)
Query 14 IEAMSNLIQAEAAKNSGLAAVENAATNRAQQAWRELTTQY 53
+E SNL+ AE N+ VEN NR ++W +T +
Sbjct 56 LEEGSNLVDAETLMNNDPFHVENVLDNRVIRSWNPVTKDF 95
> cgs:C624_09505 hypothetical protein
Length=95
Score = 32.7 bits (73), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 21/40 (53%), Gaps = 0/40 (0%)
Query 14 IEAMSNLIQAEAAKNSGLAAVENAATNRAQQAWRELTTQY 53
+E SNL+ AE N+ VEN NR ++W +T +
Sbjct 56 LEEGSNLVDAETLMNNDPFHVENVLDNRVIRSWNPVTKDF 95
> cgt:cgR_1795 hypothetical protein
Length=95
Score = 32.7 bits (73), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 21/40 (53%), Gaps = 0/40 (0%)
Query 14 IEAMSNLIQAEAAKNSGLAAVENAATNRAQQAWRELTTQY 53
+E SNL+ AE N+ VEN NR ++W +T +
Sbjct 56 LEEGSNLVDAETLMNNDPFHVENVLDNRVIRSWNPVTKDF 95
> cgu:WA5_1890 hypothetical protein
Length=95
Score = 32.7 bits (73), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 21/40 (53%), Gaps = 0/40 (0%)
Query 14 IEAMSNLIQAEAAKNSGLAAVENAATNRAQQAWRELTTQY 53
+E SNL+ AE N+ VEN NR ++W +T +
Sbjct 56 LEEGSNLVDAETLMNNDPFHVENVLDNRVIRSWNPVTKDF 95
> cgb:cg2155 hypothetical protein
Length=95
Score = 32.7 bits (73), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 21/40 (53%), Gaps = 0/40 (0%)
Query 14 IEAMSNLIQAEAAKNSGLAAVENAATNRAQQAWRELTTQY 53
+E SNL+ AE N+ VEN NR ++W +T +
Sbjct 56 LEEGSNLVDAETLMNNDPFHVENVLDNRVIRSWNPVTKDF 95
> cgl:NCgl1890 Cgl1965; hypothetical protein
Length=95
Score = 32.7 bits (73), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 21/40 (53%), Gaps = 0/40 (0%)
Query 14 IEAMSNLIQAEAAKNSGLAAVENAATNRAQQAWRELTTQY 53
+E SNL+ AE N+ VEN NR ++W +T +
Sbjct 56 LEEGSNLVDAETLMNNDPFHVENVLDNRVIRSWNPVTKDF 95
> ebt:EBL_c04600 exuR; negative regulator of exu regulon
Length=258
Score = 33.5 bits (75), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 22/53 (42%), Positives = 31/53 (58%), Gaps = 5/53 (9%)
Query 25 AAKNSGLAA-VENAATNRAQQA-WRELTTQYEYGIEKTPKFWMDLGDRILKAI 75
A +N+ LAA VE T RA W++L +E+ E+T W + DRILKA+
Sbjct 153 ATQNTALAAIVEKMWTQRAHNPYWKKL---HEHIDERTVDHWCEDHDRILKAL 202
> ngr:NAEGRDRAFT_58860 peptidase C2, calpain
Length=758
Score = 33.5 bits (75), Expect = 3.8, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 33/58 (57%), Gaps = 7/58 (12%)
Query 14 IEAMSNLIQAEAAKNSGLAAVENAATNRAQQAWRELTTQYEYGIEKTPKFWMDLGDRI 71
+ +NLIQ + +N+GL V + A++R + +E Q + G+ FWM+LGD I
Sbjct 541 VTEQANLIQ-QLCRNTGLLPVNDTASDRKSRKQKEKEEQDD-GV-----FWMELGDVI 591
> mlo:mll5522 rubredoxin reductase
Length=417
Score = 33.1 bits (74), Expect = 5.8, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 30/51 (59%), Gaps = 2/51 (4%)
Query 31 LAAVENAATNRAQQAWRELTTQYEYGIEKTPKFWMDLGDRILKAIGVTVGA 81
L +V+NA T++A+ A R +T + P FW D+GD L+ +G+T G
Sbjct 291 LESVQNA-TDQARLAARTITGHAD-AYSAVPWFWSDIGDMKLQMVGLTAGG 339
> fgi:FGOP10_02691 uracil-DNA glycosylase superfamily protein
Length=391
Score = 32.3 bits (72), Expect = 9.3, Method: Composition-based stats.
Identities = 15/32 (47%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 47 RELTTQYEYGIEKTPKFWMDLGDRILKAIGVT 78
R+L T E GI TP F+M GD++L G T
Sbjct 340 RDLKTVKELGITSTPTFFMMSGDKVLDTYGPT 371
> eus:EUTSA_v10025847mg hypothetical protein
Length=298
Score = 32.3 bits (72), Expect = 9.3, Method: Composition-based stats.
Identities = 13/22 (59%), Positives = 16/22 (73%), Gaps = 0/22 (0%)
Query 47 RELTTQYEYGIEKTPKFWMDLG 68
REL T+YEYG E P+F + LG
Sbjct 66 RELFTKYEYGAEGRPRFGISLG 87
Lambda K H a alpha
0.314 0.127 0.356 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 128026330890