bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-37_CDS_annotation_glimmer3.pl_2_1
Length=238
Score E
Sequences producing significant alignments: (Bits) Value
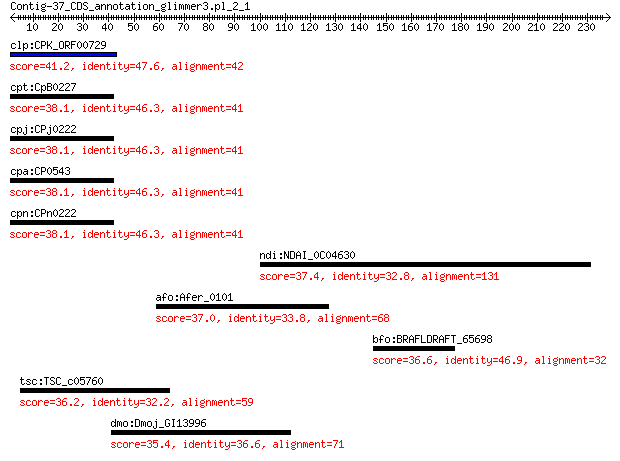
clp:CPK_ORF00729 hypothetical protein 41.2 0.031
cpt:CpB0227 hypothetical protein 38.1 0.29
cpj:CPj0222 hypothetical protein 38.1 0.29
cpa:CP0543 hypothetical protein 38.1 0.29
cpn:CPn0222 hypothetical protein 38.1 0.29
ndi:NDAI_0C04630 NDAI0C04630; hypothetical protein 37.4 3.2
afo:Afer_0101 phosphoenolpyruvate carboxylase (EC:4.1.1.31) 37.0 3.8
bfo:BRAFLDRAFT_65698 hypothetical protein 36.6 4.5
tsc:TSC_c05760 leucine-, isoleucine-, valine-, threonine-, and... 36.2 6.0
dmo:Dmoj_GI13996 GI13996 gene product from transcript GI13996-RA 35.4 7.8
> clp:CPK_ORF00729 hypothetical protein
Length=121
Score = 41.2 bits (95), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 20/42 (48%), Positives = 26/42 (62%), Gaps = 3/42 (7%)
Query 1 MQLFFKRLNQNIRSVTNEKIYYYVVGEYGPTTFRPHFHILLF 42
+QLF KRL I + KI Y+ GEYG RPH+H+L+F
Sbjct 75 LQLFLKRLRDRI---SPHKIRYFGCGEYGTKLQRPHYHLLIF 113
> cpt:CpB0227 hypothetical protein
Length=113
Score = 38.1 bits (87), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 25/41 (61%), Gaps = 3/41 (7%)
Query 1 MQLFFKRLNQNIRSVTNEKIYYYVVGEYGPTTFRPHFHILL 41
+QLF KRL + I + KI Y+ G YG RPH+H+LL
Sbjct 75 LQLFLKRLRKMI---SPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpj:CPj0222 hypothetical protein
Length=113
Score = 38.1 bits (87), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 25/41 (61%), Gaps = 3/41 (7%)
Query 1 MQLFFKRLNQNIRSVTNEKIYYYVVGEYGPTTFRPHFHILL 41
+QLF KRL + I + KI Y+ G YG RPH+H+LL
Sbjct 75 LQLFLKRLRKMI---SPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpa:CP0543 hypothetical protein
Length=113
Score = 38.1 bits (87), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 25/41 (61%), Gaps = 3/41 (7%)
Query 1 MQLFFKRLNQNIRSVTNEKIYYYVVGEYGPTTFRPHFHILL 41
+QLF KRL + I + KI Y+ G YG RPH+H+LL
Sbjct 75 LQLFLKRLRKMI---SPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpn:CPn0222 hypothetical protein
Length=113
Score = 38.1 bits (87), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 25/41 (61%), Gaps = 3/41 (7%)
Query 1 MQLFFKRLNQNIRSVTNEKIYYYVVGEYGPTTFRPHFHILL 41
+QLF KRL + I + KI Y+ G YG RPH+H+LL
Sbjct 75 LQLFLKRLRKMI---SPHKIRYFECGAYGTKLQRPHYHLLL 112
> ndi:NDAI_0C04630 NDAI0C04630; hypothetical protein
Length=3755
Score = 37.4 bits (85), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 43/137 (31%), Positives = 63/137 (46%), Gaps = 20/137 (15%)
Query 100 PFGRFSMHFAESAFNEVFKPQEDEEIFSLFYDGRMLELNGKPTLVRPKRSHINRL-YPRL 158
P S+ + NE FK DE++F L +L ++G + NRL +PR
Sbjct 3230 PLLALSLESLVAQINERFKNNTDEDLFRLI---NVLLIDGT--------FNYNRLPFPRN 3278
Query 159 NKSKHASVDDDIRVATALSNIPHVLAKFG--FIDEVTDFEM-SKRIYYLIRRYLEIDHTL 215
N +S +++ + PH+ KF FIDE DFE KR+ Y RR +++ L
Sbjct 3279 NPPLPSSTANNLARLSETLLAPHIRPKFNADFIDEKPDFETYIKRLRYWRRR---LENKL 3335
Query 216 KYAP--EQLRLIYNYLS 230
AP E L I +LS
Sbjct 3336 DRAPHVESLEKICPHLS 3352
> afo:Afer_0101 phosphoenolpyruvate carboxylase (EC:4.1.1.31)
Length=772
Score = 37.0 bits (84), Expect = 3.8, Method: Composition-based stats.
Identities = 23/68 (34%), Positives = 30/68 (44%), Gaps = 0/68 (0%)
Query 59 SWRFGDTDTQPVWSSASCYVAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAESAFNEVFK 118
SW GD D P+ ++ A N+TACL F +K SMHFA + V
Sbjct 126 SWIGGDRDGNPLVTAEVTKAAFVANATACLEAFRSRLDAVKRRLTHSMHFAAVSDAIVES 185
Query 119 PQEDEEIF 126
+ DE F
Sbjct 186 IRNDEHDF 193
> bfo:BRAFLDRAFT_65698 hypothetical protein
Length=557
Score = 36.6 bits (83), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 15/32 (47%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 145 RPKRSHINRLYPRLNKSKHASVDDDIRVATAL 176
RP SH + YPR S+ A VDDD+R T +
Sbjct 232 RPGSSHQSEAYPRPGSSRQADVDDDVRTLTEM 263
> tsc:TSC_c05760 leucine-, isoleucine-, valine-, threonine-, and
alanine-binding protein
Length=398
Score = 36.2 bits (82), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 28/59 (47%), Gaps = 7/59 (12%)
Query 5 FKRLNQNIRSVTNEKIYYYVVGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSKSWRFG 63
+R + + VTNE +Y +VG GP F+P F + +Q I +KS R G
Sbjct 314 IRRAQERFKRVTNETVYQALVGMNGPNAFKPGFAV-------STKQGIEIDFTKSERTG 365
> dmo:Dmoj_GI13996 GI13996 gene product from transcript GI13996-RA
Length=278
Score = 35.4 bits (80), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 26/80 (33%), Positives = 38/80 (48%), Gaps = 13/80 (16%)
Query 41 LFHDSKELRQ-----SIRQFVSKSWRFGDTDTQPVWSSASCYVAGYVNSTACLPDF--YK 93
L+ S +L++ SI QF+S + TD Q W+SAS +STA F +
Sbjct 174 LWQGSADLKETQYIPSIDQFISAT----TTDQQSAWNSASSMTTSACSSTASYDSFESFN 229
Query 94 NFSHIK--PFGRFSMHFAES 111
NF + PFG + +A S
Sbjct 230 NFDQLNAAPFGAQQLEWASS 249
Lambda K H a alpha
0.326 0.140 0.429 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 329549477264