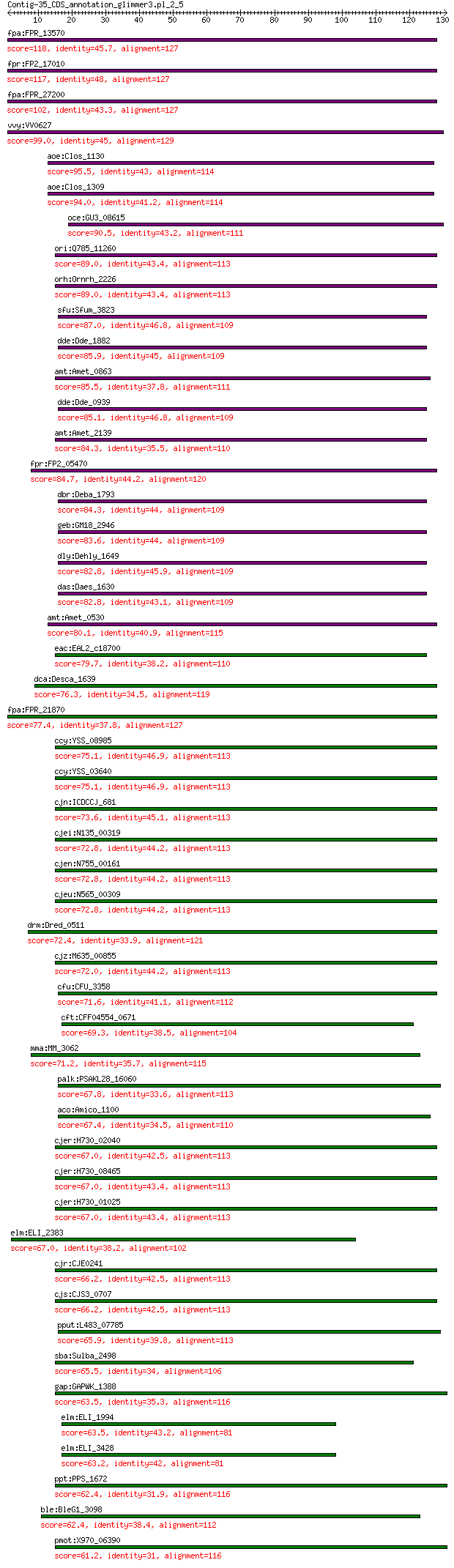
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-35_CDS_annotation_glimmer3.pl_2_5
Length=130
Score E
Sequences producing significant alignments: (Bits) Value
fpa:FPR_13570 hypothetical protein 118 1e-31
fpr:FP2_17010 hypothetical protein 117 3e-31
fpa:FPR_27200 hypothetical protein 102 4e-25
vvy:VV0627 hypothetical protein 99.0 3e-23
aoe:Clos_1130 peptidase M15A 95.5 6e-23
aoe:Clos_1309 peptidase M15A 94.0 3e-22
oce:GU3_08615 peptidase M15A 90.5 7e-21
ori:Q785_11260 peptidase M15 family protein 89.0 3e-20
orh:Ornrh_2226 hypothetical protein 89.0 3e-20
sfu:Sfum_3823 peptidase M15A 87.0 2e-19
dde:Dde_1882 peptidase M15A 85.9 3e-19
amt:Amet_0863 peptidase M15A 85.5 5e-19
dde:Dde_0939 peptidase M15A 85.1 7e-19
amt:Amet_2139 peptidase M15A 84.3 1e-18
fpr:FP2_05470 hypothetical protein 84.7 1e-18
dbr:Deba_1793 peptidase M15A 84.3 1e-18
geb:GM18_2946 peptidase M15A 83.6 3e-18
dly:Dehly_1649 peptidase M15A 82.8 5e-18
das:Daes_1630 peptidase M15A 82.8 5e-18
amt:Amet_0530 peptidase M15A 80.1 5e-17
eac:EAL2_c18700 peptidase M15A 79.7 9e-17
dca:Desca_1639 peptidase M15A 76.3 2e-15
fpa:FPR_21870 hypothetical protein 77.4 2e-15
ccy:YSS_08985 peptidase M15 family protein 75.1 5e-15
ccy:YSS_03640 peptidase M15 family protein 75.1 5e-15
cjn:ICDCCJ_681 hypothetical protein 73.6 2e-14
cjei:N135_00319 peptidase M15 family protein 72.8 4e-14
cjen:N755_00161 peptidase M15 family protein 72.8 4e-14
cjeu:N565_00309 peptidase M15 family protein 72.8 4e-14
drm:Dred_0511 peptidase M15A 72.4 4e-14
cjz:M635_00855 peptidase M15 family protein 72.0 7e-14
cfu:CFU_3358 hypothetical protein 71.6 1e-13
cft:CFF04554_0671 hypothetical protein 69.3 5e-13
mma:MM_3062 hypothetical protein 71.2 1e-12
palk:PSAKL28_16060 peptidase M15A 67.8 3e-12
aco:Amico_1100 peptidase M15A 67.4 3e-12
cjer:H730_02040 hypothetical protein 67.0 4e-12
cjer:H730_08465 hypothetical protein 67.0 5e-12
cjer:H730_01025 hypothetical protein 67.0 5e-12
elm:ELI_2383 peptidase M15A 67.0 7e-12
cjr:CJE0241 hypothetical protein 66.2 8e-12
cjs:CJS3_0707 hypothetical protein 66.2 8e-12
pput:L483_07785 peptidase M15 65.9 1e-11
sba:Sulba_2498 Peptidase M15 65.5 2e-11
gap:GAPWK_1388 hypothetical protein 63.5 1e-10
elm:ELI_1994 peptidase M15A 63.5 1e-10
elm:ELI_3428 peptidase M15A 63.2 2e-10
ppt:PPS_1672 peptidase M15A 62.4 3e-10
ble:BleG1_3098 Zinc D-Ala-D-Ala carboxypeptidase 62.4 3e-10
pmot:X970_06390 peptidase M15 61.2 6e-10
> fpa:FPR_13570 hypothetical protein
Length=137
Score = 118 bits (296), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 58/134 (43%), Positives = 81/134 (60%), Gaps = 7/134 (5%)
Query 1 MEIKAYVMKEQANEKITSHFKVKEFACKDGTPIVFVDEYLAVILDIAREKIGKPITITSG 60
M ++AY + + ++ HF V+EF CKDG+ VF+D LA +L+ RE GKP+TITS
Sbjct 1 MALQAYSLARDGSTALSPHFCVREFRCKDGSDPVFIDTALAELLERIREHFGKPVTITSA 60
Query 61 YRTVSHNAKVGGARYSFHTRGMAADIRANGIKPKELAEVLNSIVPNSCGIIVYD------ 114
YRT +HNAK GGA++S H G AADIR + +++A S++P+ G+ Y
Sbjct 61 YRTPAHNAKAGGAKFSQHLYGRAADIRVQDVSVEDVAAYAESLMPDRGGVGRYPAKAGRA 120
Query 115 -NWVHFDTRNTKYR 127
WVH DTR K R
Sbjct 121 AGWVHVDTRADKAR 134
> fpr:FP2_17010 hypothetical protein
Length=137
Score = 117 bits (294), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 61/134 (46%), Positives = 80/134 (60%), Gaps = 7/134 (5%)
Query 1 MEIKAYVMKEQANEKITSHFKVKEFACKDGTPIVFVDEYLAVILDIAREKIGKPITITSG 60
M IKAY + + N+K++++F VKEF CKDGT +F+D+ L +L R GK +TITS
Sbjct 1 MAIKAYSLAKDGNKKLSANFAVKEFRCKDGTDPIFIDDVLVKLLQNIRNHFGKAVTITSA 60
Query 61 YRTVSHNAKVGGARYSFHTRGMAADIRANGIKPKELAEVLNSIVPNSCGIIVYD------ 114
YRT +HN V GA YS H GMAADIR G+ + LA +++ N+ GI Y
Sbjct 61 YRTAAHNKAVKGATYSQHCYGMAADIRIQGVDVETLATYAETLLKNTGGIGRYPVKTGRP 120
Query 115 -NWVHFDTRNTKYR 127
WVH DTR K R
Sbjct 121 AGWVHIDTRAVKSR 134
> fpa:FPR_27200 hypothetical protein
Length=140
Score = 102 bits (253), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 55/137 (40%), Positives = 78/137 (57%), Gaps = 10/137 (7%)
Query 1 MEIKAYVMKEQANEKITSHFKVKEFACKDGTPIVFVDEYLAVILDIAREKIGKPITITSG 60
M + Y ++ + ++ +FKVKEF CKDG+ +F+D L IL R+ G P+ I S
Sbjct 1 MALNVYSLERDGEKSLSKNFKVKEFRCKDGSDPIFIDSELVRILQKVRDHFGSPVIINSA 60
Query 61 YRTVSHN--AKVGGARYSFHTRGMAADIRANGIKPKELAEVLNSIVPNSCGIIVYD---- 114
YRT ++N KVGGA++S H G AADI GI +LAE + +++PN GI +Y
Sbjct 61 YRTAAYNLSKKVGGAKFSQHQYGKAADIYIQGILITKLAEYVETLMPNKGGIGIYPIKTG 120
Query 115 ----NWVHFDTRNTKYR 127
+VH D R TK R
Sbjct 121 VRNCAFVHVDVRATKGR 137
> vvy:VV0627 hypothetical protein
Length=227
Score = 99.0 bits (245), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 58/143 (41%), Positives = 89/143 (62%), Gaps = 15/143 (10%)
Query 1 MEIKAYVMKEQANEK---ITSHFKVKEFAC--KDGTPIVFVD-----EYLAVILDIAREK 50
+++ + ++K +A++ +T+HFK EF+C KD TP V D + LA L++ R +
Sbjct 85 VKLSSAILKLKAHQNSSILTAHFKWSEFSCNDKDNTP-VPQDLRTNVKNLASQLEVIRSE 143
Query 51 IGKPITITSGYRTVSHNAKVGGARYSFHTRGMAADIRANGIKPKELAEVLNSIVPN---- 106
IGKPI ITS YRT +N K+GGA S H G AAD++ +G+KPK+L E + S++ N
Sbjct 144 IGKPIKITSAYRTPEYNRKIGGATNSLHVTGKAADLQVSGVKPKDLYEKIISLINNGKIT 203
Query 107 SCGIIVYDNWVHFDTRNTKYRKG 129
G+ +Y ++VH+D R T R G
Sbjct 204 QGGVGLYTSFVHYDIRGTSARWG 226
> aoe:Clos_1130 peptidase M15A
Length=120
Score = 95.5 bits (236), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 49/114 (43%), Positives = 68/114 (60%), Gaps = 2/114 (2%)
Query 13 NEKITSHFKVKEFACKDGTPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVGG 72
N +I+ +FK+KEF C G +V +D L L + R K+ PI ITSGYRT+ N +VGG
Sbjct 3 NIQISKNFKLKEFECSHGGSVVKLDSKLLEKLQLLRVKLNNPINITSGYRTLECNKRVGG 62
Query 73 ARYSFHTRGMAADIRANGIKPKELAEVLNSIVPNSCGIIVYDNWVHFDTRNTKY 126
+ S+H +GMAADI + G P ++A+ + GI Y N+VH D R KY
Sbjct 63 SSNSYHMKGMAADIYSPGYTPTQIAKAAEEV--GFTGIGTYSNFVHVDVRPNKY 114
> aoe:Clos_1309 peptidase M15A
Length=120
Score = 94.0 bits (232), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 47/114 (41%), Positives = 68/114 (60%), Gaps = 2/114 (2%)
Query 13 NEKITSHFKVKEFACKDGTPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVGG 72
N +I+ +FK+KEF C G +V +D L L + R+K+ PI +TSGYRT N +VGG
Sbjct 3 NIQISKNFKLKEFDCSHGDSVVKLDSRLLEKLQLLRDKLNNPINVTSGYRTPECNKRVGG 62
Query 73 ARYSFHTRGMAADIRANGIKPKELAEVLNSIVPNSCGIIVYDNWVHFDTRNTKY 126
+ S+H +GMAADI + G P ++A+ + GI +Y +VH D R KY
Sbjct 63 SSNSYHMKGMAADIYSPGYTPAQIAKAAEEV--GFTGIGIYSTFVHVDVRPNKY 114
> oce:GU3_08615 peptidase M15A
Length=123
Score = 90.5 bits (223), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 48/112 (43%), Positives = 64/112 (57%), Gaps = 1/112 (1%)
Query 19 HFKVKEFACKDGTPIVFVDEYLAVILDIAREKIG-KPITITSGYRTVSHNAKVGGARYSF 77
HF +EFACK G VD L +L+ R+ +G +P++ITS R HN KVGGAR+S
Sbjct 10 HFARREFACKCGCGFDAVDIELLGVLEQLRQDLGNRPVSITSACRCEPHNRKVGGARHSV 69
Query 78 HTRGMAADIRANGIKPKELAEVLNSIVPNSCGIIVYDNWVHFDTRNTKYRKG 129
H +G AADIR G+ P ++A+ L P+ GI Y + H D R R G
Sbjct 70 HKQGKAADIRVKGLLPAQVADYLERTYPDRYGIGRYATFTHIDVRPAPARWG 121
> ori:Q785_11260 peptidase M15 family protein
Length=125
Score = 89.0 bits (219), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 49/121 (40%), Positives = 69/121 (57%), Gaps = 8/121 (7%)
Query 15 KITSHFKVKEFACKDG--TPIVFVDEY--LAVILDIAREKIGKPITITSGYRTVSHNAKV 70
K++ F +KEF C DG TP + LA L R +GKPI + SGYR+V+HN K+
Sbjct 2 KLSKDFDLKEFECHDGSTTPAFALKNLRELASNLQELRNYLGKPIIVNSGYRSVAHNRKI 61
Query 71 GGARYSFHTRGMAADIRANGIKPKELAEVLNSIVPN----SCGIIVYDNWVHFDTRNTKY 126
GG S H +G AADIR G+ P++LA + ++ N G+ +Y +VH+D R K
Sbjct 62 GGKVNSQHLQGKAADIRVEGVTPEKLASTIERLIFNGKMKQGGLGIYLTFVHYDIRGIKA 121
Query 127 R 127
R
Sbjct 122 R 122
> orh:Ornrh_2226 hypothetical protein
Length=125
Score = 89.0 bits (219), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 49/121 (40%), Positives = 69/121 (57%), Gaps = 8/121 (7%)
Query 15 KITSHFKVKEFACKDG--TPIVFVDEY--LAVILDIAREKIGKPITITSGYRTVSHNAKV 70
K++ F +KEF C DG TP + LA L R +GKPI + SGYR+V+HN K+
Sbjct 2 KLSKDFDLKEFECHDGSTTPAFALKNLRELASNLQELRNYLGKPIIVNSGYRSVAHNRKI 61
Query 71 GGARYSFHTRGMAADIRANGIKPKELAEVLNSIVPN----SCGIIVYDNWVHFDTRNTKY 126
GG S H +G AADIR G+ P++LA + ++ N G+ +Y +VH+D R K
Sbjct 62 GGKVNSQHLQGKAADIRVEGVTPEKLASTIERLIFNGKMKQGGLGIYLTFVHYDIRGIKA 121
Query 127 R 127
R
Sbjct 122 R 122
> sfu:Sfum_3823 peptidase M15A
Length=124
Score = 87.0 bits (214), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 51/115 (44%), Positives = 67/115 (58%), Gaps = 7/115 (6%)
Query 16 ITSHFKVKEFACKDGTPIV----FVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVG 71
++ +F EFACK GT V L L R++IGKP++ITSG+R HN VG
Sbjct 4 LSKNFNRSEFACK-GTNCCGHSAAVHPDLVDALQTLRDRIGKPLSITSGFRCNRHNKAVG 62
Query 72 GARYSFHTRGMAADIRA-NGIKPKELAEVLNSI-VPNSCGIIVYDNWVHFDTRNT 124
GA SFHT GMAAD+ G+ P+ELA + I + + GI VY +WVH D R +
Sbjct 63 GAEQSFHTLGMAADVSCPAGVSPEELAVIAEEIPLFHEGGIGVYASWVHLDVRQS 117
> dde:Dde_1882 peptidase M15A
Length=124
Score = 85.9 bits (211), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 49/114 (43%), Positives = 64/114 (56%), Gaps = 5/114 (4%)
Query 16 ITSHFKVKEFACKDGTPI---VFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVGG 72
++ +F EFACK V L L R+++GKP++ITSG+R HN VGG
Sbjct 4 LSKNFNRSEFACKGKNCCGHSAAVHPDLVDALQALRDRVGKPLSITSGFRCNRHNKAVGG 63
Query 73 ARYSFHTRGMAADIRA-NGIKPKELAEVLNSI-VPNSCGIIVYDNWVHFDTRNT 124
A SFHT GMAAD+ G+ P+ELA V I + GI VY +WVH D R +
Sbjct 64 AEQSFHTLGMAADVSCPAGVSPEELAVVAEEIPLFREGGIGVYASWVHLDVRQS 117
> amt:Amet_0863 peptidase M15A
Length=119
Score = 85.5 bits (210), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 42/111 (38%), Positives = 64/111 (58%), Gaps = 2/111 (2%)
Query 15 KITSHFKVKEFACKDGTPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVGGAR 74
+I +F+++EF C+ G+ +V +D L L R ++ PI +TSGYRT HN +VGG+
Sbjct 5 QIAKNFQLREFQCRGGSQLVKLDHQLLEKLQQLRNQVNAPINLTSGYRTPEHNQRVGGSP 64
Query 75 YSFHTRGMAADIRANGIKPKELAEVLNSIVPNSCGIIVYDNWVHFDTRNTK 125
S H G AADI+ G P+ +A++ I GI +Y + H D R T+
Sbjct 65 NSQHLLGRAADIQVPGHSPEAIAKMAEKI--GFAGIGIYSTFTHVDVRTTE 113
> dde:Dde_0939 peptidase M15A
Length=124
Score = 85.1 bits (209), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 51/115 (44%), Positives = 65/115 (57%), Gaps = 7/115 (6%)
Query 16 ITSHFKVKEFACKDGTPIV----FVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVG 71
++ +F EFACK GT V L L R+ IGKP++ITSG+R HN VG
Sbjct 4 LSKNFNRSEFACK-GTNCCGHSAAVHPDLVDALQALRDHIGKPLSITSGFRCNRHNKAVG 62
Query 72 GARYSFHTRGMAADIRA-NGIKPKELAEVLNSI-VPNSCGIIVYDNWVHFDTRNT 124
GA SFHT GMAAD+ G+ P+ELA + I + GI VY +WVH D R +
Sbjct 63 GAEQSFHTLGMAADVSCPAGVSPEELAVIAEEIPLFREGGIGVYASWVHLDVRQS 117
> amt:Amet_2139 peptidase M15A
Length=119
Score = 84.3 bits (207), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 39/110 (35%), Positives = 66/110 (60%), Gaps = 2/110 (2%)
Query 15 KITSHFKVKEFACKDGTPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVGGAR 74
+I +F+++EF C G+ +V +D L L R+++G P+ +TSG+RT HN +VGG+
Sbjct 5 QIDKNFQLREFQCLGGSQLVKLDHRLIEKLQQLRDQVGSPVIVTSGFRTPEHNKRVGGSL 64
Query 75 YSFHTRGMAADIRANGIKPKELAEVLNSIVPNSCGIIVYDNWVHFDTRNT 124
S H G AADI+ G P+ +A++ +++ G+ +Y + H D R T
Sbjct 65 NSQHLLGRAADIQVPGYSPEAIAQIADAL--GFTGVGIYATFTHVDVRTT 112
> fpr:FP2_05470 hypothetical protein
Length=132
Score = 84.7 bits (208), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 53/130 (41%), Positives = 74/130 (57%), Gaps = 11/130 (8%)
Query 8 MKEQANEKITSHFKVKEFACKDGTPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHN 67
M + +++ +FKV+EFACK G+ +V +D+ L V+L RE GKP+ ITSGYRT +HN
Sbjct 1 MSRDSTRQLSPNFKVREFACK-GSDVVLLDDELVVLLQCIREHFGKPVHITSGYRTAAHN 59
Query 68 AKVGGARYSFHTRGMAADIRANGIKPKELAEVLNSIVPNSCGIIVYD----------NWV 117
A VGG++ S H G AAD G+ +A +++P+ GI Y WV
Sbjct 60 AAVGGSKSSQHLLGRAADFYVEGVDVATVAAYAETLLPSRGGIGRYPKDAAHPKRRTGWV 119
Query 118 HFDTRNTKYR 127
H DTR K R
Sbjct 120 HIDTRAGKSR 129
> dbr:Deba_1793 peptidase M15A
Length=124
Score = 84.3 bits (207), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 48/114 (42%), Positives = 64/114 (56%), Gaps = 5/114 (4%)
Query 16 ITSHFKVKEFACKDGTPI---VFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVGG 72
++ +F EFACK V L L R++IGKP++ITSG+R HN VGG
Sbjct 4 LSKNFNRSEFACKGKNCCGHSAAVHPDLVDALQALRDRIGKPLSITSGFRCNRHNKAVGG 63
Query 73 ARYSFHTRGMAADIRA-NGIKPKELAEVLNSI-VPNSCGIIVYDNWVHFDTRNT 124
A SFHT GMAAD+ G+ P++LA + I + GI VY +WVH D R +
Sbjct 64 AEQSFHTLGMAADVSCPAGVSPEQLAVIAEEIPLFREGGIGVYASWVHLDVRRS 117
> geb:GM18_2946 peptidase M15A
Length=124
Score = 83.6 bits (205), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 48/114 (42%), Positives = 63/114 (55%), Gaps = 5/114 (4%)
Query 16 ITSHFKVKEFACKDGTPI---VFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVGG 72
++ +F EFACK V L L R++IGKP++ITSG+R HN VGG
Sbjct 4 LSKNFNRSEFACKGKNCCGHSAAVHPDLVDALQTLRDRIGKPLSITSGFRCNRHNKAVGG 63
Query 73 ARYSFHTRGMAADIRA-NGIKPKELAEVLNSI-VPNSCGIIVYDNWVHFDTRNT 124
A SFHT GMAAD+ G+ P+ LA + I + GI VY +WVH D R +
Sbjct 64 AEQSFHTLGMAADVSCPAGVSPEALAVIAEEIPLFREGGIGVYASWVHLDVRQS 117
> dly:Dehly_1649 peptidase M15A
Length=124
Score = 82.8 bits (203), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 50/115 (43%), Positives = 64/115 (56%), Gaps = 7/115 (6%)
Query 16 ITSHFKVKEFACKDGTPIV----FVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVG 71
++ +F EFACK GT V L L R++IGKP++ITSG+R HN VG
Sbjct 4 LSKNFNRSEFACK-GTNCCGHSAAVHPDLVDALQTLRDRIGKPLSITSGFRCNRHNKAVG 62
Query 72 GARYSFHTRGMAADIRA-NGIKPKELAEVLNSI-VPNSCGIIVYDNWVHFDTRNT 124
GA SFHT GMAAD+ G+ P LA + I + GI VY +WVH D R +
Sbjct 63 GAEKSFHTLGMAADVSCPAGVSPDALAVIAEEIPLFREGGIGVYASWVHLDVRQS 117
> das:Daes_1630 peptidase M15A
Length=124
Score = 82.8 bits (203), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 47/114 (41%), Positives = 64/114 (56%), Gaps = 5/114 (4%)
Query 16 ITSHFKVKEFACKDGTPI---VFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVGG 72
++ +F EFACK V L L R++IGKP++ITSG+R HN VGG
Sbjct 4 LSKNFNRSEFACKGKNCCGHSAAVHPDLVDALQALRDRIGKPLSITSGFRCNRHNKAVGG 63
Query 73 ARYSFHTRGMAADIRA-NGIKPKELAEVLNSI-VPNSCGIIVYDNWVHFDTRNT 124
A S+HT GMAAD+ +G+ P +LA + I + GI VY +WVH D R +
Sbjct 64 AAQSYHTLGMAADVSCPDGVSPGDLAVIAEEIPLFREGGIGVYASWVHLDVRQS 117
> amt:Amet_0530 peptidase M15A
Length=118
Score = 80.1 bits (196), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 47/115 (41%), Positives = 66/115 (57%), Gaps = 2/115 (2%)
Query 13 NEKITSHFKVKEFACKDGTPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVGG 72
N +I+ +FK+KEF C DG V +D + L RE+ G+P+ I SGYRT S+N +VGG
Sbjct 3 NIQISKNFKLKEFQCGDGGYQVRLDSQVLKKLQELREQTGRPVLINSGYRTPSYNQQVGG 62
Query 73 ARYSFHTRGMAADIRANGIKPKELAEVLNSIVPNSCGIIVYDNWVHFDTRNTKYR 127
+ S H G AADI G++ + LA V I I +Y ++H D R+ K R
Sbjct 63 SPRSQHLLGKAADIMVPGMELESLARVAEGIGFGG--IGIYRTFIHVDVRSEKVR 115
> eac:EAL2_c18700 peptidase M15A
Length=130
Score = 79.7 bits (195), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 42/110 (38%), Positives = 63/110 (57%), Gaps = 2/110 (2%)
Query 15 KITSHFKVKEFACKDGTPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVGGAR 74
K++ F + EFAC+D V+VD L L + R+ +P+ ITSGYR+ +N VGGA
Sbjct 8 KLSRDFSLSEFACRDEKRSVYVDAELVQRLQLLRDYFKRPVVITSGYRSREYNRSVGGAD 67
Query 75 YSFHTRGMAADIRANGIKPKELAEVLNSIVPNSCGIIVYDNWVHFDTRNT 124
S H G AADI+ G+ +++A +I G+ +Y ++VH D R T
Sbjct 68 GSQHLEGRAADIKVIGVGMEDIARA--AIQAGFKGVGIYGSFVHVDVRKT 115
> dca:Desca_1639 peptidase M15A
Length=128
Score = 76.3 bits (186), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 41/119 (34%), Positives = 62/119 (52%), Gaps = 1/119 (1%)
Query 9 KEQANEKITSHFKVKEFACKDGTPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNA 68
K + ++IT HF E AC+ ++ E L L+ R+ + KP+ + SGYR +HN
Sbjct 6 KAKPEQRITPHFTEGELACRCCGRLLVQSE-LVHKLETLRQLVKKPVLVNSGYRCPAHNR 64
Query 69 KVGGARYSFHTRGMAADIRANGIKPKELAEVLNSIVPNSCGIIVYDNWVHFDTRNTKYR 127
VGGA S+H +GMAADI G+ EL+ + N G +++H D R + R
Sbjct 65 AVGGAVNSYHLKGMAADIHVPGLAVVELSRLAEQAGFNGIGTYPKQSFLHVDVRGNRAR 123
> fpa:FPR_21870 hypothetical protein
Length=215
Score = 77.4 bits (189), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 48/135 (36%), Positives = 67/135 (50%), Gaps = 8/135 (6%)
Query 1 MEIKAYVMKEQANEKITSHFKVKEFACKDGTPIVFVDEYLAVILDIAREKIGKPITITSG 60
M + Y MK+ N+K++ +F EFAC D + V L L R+ GKP+ I+S
Sbjct 1 MSVITYSMKKDWNKKLSKNFCAYEFACNDRSDEFKVATELVETLQQIRDHFGKPVLISSA 60
Query 61 YRTVSHNAKVGGARYSFHTRGMAADIRANGIKPKELAEVLNSI--VPNSCGIIVYD---- 114
YRT ++N +GG+ S H G AADI NG+ P +A + S+ GI Y
Sbjct 61 YRTPAYNISIGGSSRSQHCLGTAADIHINGVDPIRIALYVASLPYFQKHGGIGYYSRAQV 120
Query 115 --NWVHFDTRNTKYR 127
+VH D R T R
Sbjct 121 TGGFVHIDVRETHSR 135
> ccy:YSS_08985 peptidase M15 family protein
Length=129
Score = 75.1 bits (183), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 53/125 (42%), Positives = 65/125 (52%), Gaps = 13/125 (10%)
Query 15 KITSHFKVKEFACKDG----TPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKV 70
K +FK EF CK G V DE + ++ +I RE PI I SGYR HNAKV
Sbjct 2 KNNPYFKESEFKCKCGKCELPQNVPSDELIDILCEI-REYYNAPIIINSGYRCKEHNAKV 60
Query 71 GGARYSFHTRGMAADIRANGIKPKELAE-VLNSIVPNSCGIIV-------YDNWVHFDTR 122
GGA S HT G AAD G+K +E+ + VLN+ S GI + Y +VH DTR
Sbjct 61 GGAPKSQHTIGSAADFVVKGVKTEEVHQYVLNTYGERSLGIAIKHNFNDPYAGFVHLDTR 120
Query 123 NTKYR 127
K R
Sbjct 121 GKKAR 125
> ccy:YSS_03640 peptidase M15 family protein
Length=129
Score = 75.1 bits (183), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 53/125 (42%), Positives = 65/125 (52%), Gaps = 13/125 (10%)
Query 15 KITSHFKVKEFACKDG----TPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKV 70
K +FK EF CK G V DE + ++ +I RE PI I SGYR HNAKV
Sbjct 2 KNNPYFKESEFKCKCGKCELPQNVPSDELIDILCEI-REYYNAPIIINSGYRCKEHNAKV 60
Query 71 GGARYSFHTRGMAADIRANGIKPKELAE-VLNSIVPNSCGIIV-------YDNWVHFDTR 122
GGA S HT G AAD G+K +E+ + VLN+ S GI + Y +VH DTR
Sbjct 61 GGAPKSQHTIGSAADFVVKGVKTEEVHQYVLNTYGERSLGIAIKHNFNDPYAGFVHLDTR 120
Query 123 NTKYR 127
K R
Sbjct 121 GKKAR 125
> cjn:ICDCCJ_681 hypothetical protein
Length=129
Score = 73.6 bits (179), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 51/125 (41%), Positives = 65/125 (52%), Gaps = 13/125 (10%)
Query 15 KITSHFKVKEFACKDG----TPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKV 70
K +FK EF CK G V DE + ++ +I RE PI I SGYR HNA++
Sbjct 2 KNNPYFKESEFKCKCGKCELPQNVPSDELIDILCEI-REHYNAPIIINSGYRCKEHNAEI 60
Query 71 GGARYSFHTRGMAADIRANGIKPKELAE-VLNSIVPNSCGIIV-------YDNWVHFDTR 122
GGA S HT G AAD G+K +E+ + VLN+ S GI + Y +VH DTR
Sbjct 61 GGAPKSQHTIGSAADFVVKGVKTEEVHQYVLNTYGERSLGIAIKHNFNDPYAGFVHLDTR 120
Query 123 NTKYR 127
K R
Sbjct 121 GKKAR 125
> cjei:N135_00319 peptidase M15 family protein
Length=129
Score = 72.8 bits (177), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 50/125 (40%), Positives = 64/125 (51%), Gaps = 13/125 (10%)
Query 15 KITSHFKVKEFACKDG----TPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKV 70
K +FK EF CK G V DE + ++ +I RE PI I SGYR HNA++
Sbjct 2 KNNPYFKESEFKCKCGKCELPQNVPSDELIDILCEI-REHYNAPIIINSGYRCKEHNAEI 60
Query 71 GGARYSFHTRGMAADIRANGIKPKELAE-VLNSIVPNSCGIIV-------YDNWVHFDTR 122
GGA S HT G AAD G+K +E+ + VLN+ GI + Y +VH DTR
Sbjct 61 GGAPKSQHTLGSAADFVVKGVKTEEVHQYVLNTYGERGLGIAIKHNFNDPYAGFVHLDTR 120
Query 123 NTKYR 127
K R
Sbjct 121 GKKAR 125
> cjen:N755_00161 peptidase M15 family protein
Length=129
Score = 72.8 bits (177), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 50/125 (40%), Positives = 64/125 (51%), Gaps = 13/125 (10%)
Query 15 KITSHFKVKEFACKDG----TPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKV 70
K +FK EF CK G V DE + ++ +I RE PI I SGYR HNA++
Sbjct 2 KNNPYFKESEFKCKCGKCELPQNVPSDELIDILCEI-REHYNAPIIINSGYRCKEHNAEI 60
Query 71 GGARYSFHTRGMAADIRANGIKPKELAE-VLNSIVPNSCGIIV-------YDNWVHFDTR 122
GGA S HT G AAD G+K +E+ + VLN+ GI + Y +VH DTR
Sbjct 61 GGAPKSQHTLGSAADFVVKGVKTEEVHQYVLNTYGERGLGIAIKHNFNDPYAGFVHLDTR 120
Query 123 NTKYR 127
K R
Sbjct 121 GKKAR 125
> cjeu:N565_00309 peptidase M15 family protein
Length=129
Score = 72.8 bits (177), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 50/125 (40%), Positives = 64/125 (51%), Gaps = 13/125 (10%)
Query 15 KITSHFKVKEFACKDG----TPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKV 70
K +FK EF CK G V DE + ++ +I RE PI I SGYR HNA++
Sbjct 2 KNNPYFKESEFKCKCGKCELPQNVPSDELIDILCEI-REHYNAPIIINSGYRCKEHNAEI 60
Query 71 GGARYSFHTRGMAADIRANGIKPKELAE-VLNSIVPNSCGIIV-------YDNWVHFDTR 122
GGA S HT G AAD G+K +E+ + VLN+ GI + Y +VH DTR
Sbjct 61 GGAPKSQHTLGSAADFVVKGVKTEEVHQYVLNTYGERGLGIAIKHNFNDPYAGFVHLDTR 120
Query 123 NTKYR 127
K R
Sbjct 121 GKKAR 125
> drm:Dred_0511 peptidase M15A
Length=124
Score = 72.4 bits (176), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 41/121 (34%), Positives = 67/121 (55%), Gaps = 3/121 (2%)
Query 7 VMKEQANEKITSHFKVKEFACKDGTPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSH 66
+ K + + +++HF E AC+ +V E + + D+ R + KP+ + SGYR ++
Sbjct 4 IKKAREGQYLSAHFAETELACRCCGKLVIHLELVYKLEDL-RRLLDKPVLVNSGYRCPTN 62
Query 67 NAKVGGARYSFHTRGMAADIRANGIKPKELAEVLNSIVPNSCGIIVYDNWVHFDTRNTKY 126
N VGG SFH++GMAADIR + KE+A + + GI +Y + VH D R+ +
Sbjct 63 NRAVGGVVNSFHSKGMAADIRVPRMAVKEIAHLAEKV--GFGGIGIYASQVHVDVRDYRT 120
Query 127 R 127
R
Sbjct 121 R 121
> cjz:M635_00855 peptidase M15 family protein
Length=129
Score = 72.0 bits (175), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 50/125 (40%), Positives = 64/125 (51%), Gaps = 13/125 (10%)
Query 15 KITSHFKVKEFACKDG----TPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKV 70
K +FK EF CK G V DE + ++ +I RE PI I SGYR HNA++
Sbjct 2 KNNPYFKESEFKCKCGKCELPQNVPSDELIDILCEI-REHYNAPIIINSGYRCKEHNAEI 60
Query 71 GGARYSFHTRGMAADIRANGIKPKELAE-VLNSIVPNSCGIIV-------YDNWVHFDTR 122
GGA S HT G AAD G+K +E+ + VLN+ GI + Y +VH DTR
Sbjct 61 GGAPKSQHTIGSAADFVVKGVKTEEVHQYVLNTYGERGLGIAIKHNFNDPYAGFVHLDTR 120
Query 123 NTKYR 127
K R
Sbjct 121 GKKAR 125
> cfu:CFU_3358 hypothetical protein
Length=143
Score = 71.6 bits (174), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 46/124 (37%), Positives = 62/124 (50%), Gaps = 12/124 (10%)
Query 16 ITSHFKVKEFACK--------DGTPIVFVDEYL---AVILDIAREKIGKPITITSGYRTV 64
+T HF + E D TP + V + L A L+ R +GKPI +TSGYR+
Sbjct 3 LTEHFTLAEMTISETAVRQGIDNTPSLAVIDNLKLTAATLEQIRTLVGKPIIVTSGYRSP 62
Query 65 SHNAKVGGARYSFHTRGMAADIRANGIKPKELAEVL-NSIVPNSCGIIVYDNWVHFDTRN 123
+ N VGGA S H G+AADI G PK LA ++ +S + I+ +D WVH
Sbjct 63 AVNKAVGGASNSAHVLGLAADINVPGYTPKALANLIKDSGIQFDQLILEFDRWVHVGLAA 122
Query 124 TKYR 127
K R
Sbjct 123 GKLR 126
> cft:CFF04554_0671 hypothetical protein
Length=121
Score = 69.3 bits (168), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 40/108 (37%), Positives = 57/108 (53%), Gaps = 7/108 (6%)
Query 17 TSHFKVKEFACKDGTPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVGGARYS 76
+F +E CK D++LA++ DI R+K P I S YR HN VGGA YS
Sbjct 4 NCYFTDEELKCKHCNKNYINDDFLALLCDI-RKKANIPFIINSAYRCKEHNKAVGGANYS 62
Query 77 FHTRGMAADIRANGIKPKELAEVLNSIVP----NSCGIIVYDNWVHFD 120
H +G+A DI+ EL +++ SI+ N I++Y +VHFD
Sbjct 63 KHVKGLAVDIKYKD--SVELYKIVKSILDFNTLNKSRILIYKTFVHFD 108
> mma:MM_3062 hypothetical protein
Length=351
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 41/115 (36%), Positives = 59/115 (51%), Gaps = 0/115 (0%)
Query 8 MKEQANEKITSHFKVKEFACKDGTPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHN 67
++++ E I+ +FKVKE A DG + L L R+++G + + SGYR N
Sbjct 45 VRDRMQENISKNFKVKELAATDGARYARISPELVAGLQRIRDRVGSAVVLNSGYRHNVLN 104
Query 68 AKVGGARYSFHTRGMAADIRANGIKPKELAEVLNSIVPNSCGIIVYDNWVHFDTR 122
VGGA S H G AADIRA+ P +LA + + GI + N +H D R
Sbjct 105 ETVGGADESQHITGRAADIRASAKSPLDLARIALEELGCDIGIGLGRNSIHVDLR 159
> palk:PSAKL28_16060 peptidase M15A
Length=143
Score = 67.8 bits (164), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 38/125 (30%), Positives = 61/125 (49%), Gaps = 12/125 (10%)
Query 16 ITSHFKVKEFACKDGTPIVFVD-----------EYLAVILDIAREKIGKPITITSGYRTV 64
++ HF + E + + +D L L+ R+ + KP+ ++SGYR +
Sbjct 3 LSEHFTLAEMTVSETAARLGIDNTPDAQTLVNLRRLCTFLEQIRQLVDKPVLVSSGYRCL 62
Query 65 SHNAKVGGARYSFHTRGMAADIRANGIKPKELAE-VLNSIVPNSCGIIVYDNWVHFDTRN 123
N +GG+ S HT G+AADI G+ P+ELA V +S + I+ +D WVH
Sbjct 63 QLNEVIGGSLQSAHTDGLAADINVPGLTPRELARGVADSSLMFDQLILEFDQWVHVGLSL 122
Query 124 TKYRK 128
T R+
Sbjct 123 TPERR 127
> aco:Amico_1100 peptidase M15A
Length=125
Score = 67.4 bits (163), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 38/115 (33%), Positives = 57/115 (50%), Gaps = 5/115 (4%)
Query 16 ITSHFKVKEFACKDGTPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVGGARY 75
++ HF E AC+ G + L +L+ R +G PI I SGYR +HN ++GG
Sbjct 2 LSEHFSRTELACRCGCDRCDIKPKLLSLLEKIRSLVGTPIFINSGYRCPTHNKRIGGVPN 61
Query 76 SFHTRGMAADIRA-----NGIKPKELAEVLNSIVPNSCGIIVYDNWVHFDTRNTK 125
S+HT+G+AADIR N K L + + G+ +Y+ +H D K
Sbjct 62 SWHTQGVAADIRQAKYSNNVFHSKVLRAYKDGKLSELGGLGLYNGRIHVDVHKPK 116
> cjer:H730_02040 hypothetical protein
Length=129
Score = 67.0 bits (162), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 48/125 (38%), Positives = 62/125 (50%), Gaps = 13/125 (10%)
Query 15 KITSHFKVKEFACKDG----TPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKV 70
K +FK EF CK G V DE + ++ +I RE P+ I SGYR HNA+V
Sbjct 2 KNNPYFKESEFKCKCGKCELPQNVPSDELIDILCEI-REHYNAPVIINSGYRCKEHNAEV 60
Query 71 GGARYSFHTRGMAADIRANGIKPKELAE-VLNSIVPNSCGIIV-------YDNWVHFDTR 122
GGA S H G AAD G+K +E+ + VL + GI + Y +VH DTR
Sbjct 61 GGAPKSQHAIGSAADFMVKGVKTEEVHQYVLITYGERGLGIAIKHNFNDPYAGFVHLDTR 120
Query 123 NTKYR 127
K R
Sbjct 121 GKKAR 125
> cjer:H730_08465 hypothetical protein
Length=129
Score = 67.0 bits (162), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 49/125 (39%), Positives = 62/125 (50%), Gaps = 13/125 (10%)
Query 15 KITSHFKVKEFACKDG----TPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKV 70
K +FK EF CK G V DE + ++ +I RE PI I SGYR HNA+V
Sbjct 2 KNNPYFKESEFKCKCGKCELPQNVPSDELIDILCEI-REHYNAPIIINSGYRCKEHNAEV 60
Query 71 GGARYSFHTRGMAADIRANGIKPKELAE-VLNSIVPNSCGIIV-------YDNWVHFDTR 122
GGA S H G AAD G+K +E+ + VL + GI + Y +VH DTR
Sbjct 61 GGAPKSQHAIGSAADFVVKGVKTEEVHQYVLITYGERGLGIAIKHNFNDPYAGFVHLDTR 120
Query 123 NTKYR 127
K R
Sbjct 121 GKKAR 125
> cjer:H730_01025 hypothetical protein
Length=129
Score = 67.0 bits (162), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 49/125 (39%), Positives = 62/125 (50%), Gaps = 13/125 (10%)
Query 15 KITSHFKVKEFACKDG----TPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKV 70
K +FK EF CK G V DE + ++ +I RE PI I SGYR HNA+V
Sbjct 2 KNNPYFKESEFKCKCGKCELPQNVPSDELIDILCEI-REHYNAPIIINSGYRCKEHNAEV 60
Query 71 GGARYSFHTRGMAADIRANGIKPKELAE-VLNSIVPNSCGIIV-------YDNWVHFDTR 122
GGA S H G AAD G+K +E+ + VL + GI + Y +VH DTR
Sbjct 61 GGAPKSQHAIGSAADFVVKGVKTEEVHQYVLITYGERGLGIAIKHNFNDPYAGFVHLDTR 120
Query 123 NTKYR 127
K R
Sbjct 121 GKKAR 125
> elm:ELI_2383 peptidase M15A
Length=186
Score = 67.0 bits (162), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 39/107 (36%), Positives = 56/107 (52%), Gaps = 6/107 (6%)
Query 2 EIKAYVMKEQANEKITSHFKVKEF-----ACKDGTPIVFVDEYLAVILDIAREKIGKPIT 56
E KA +++ +SHF ++E+ C DG P E L I + R G+P+
Sbjct 62 EPKAPEPEQREPLMASSHFAMEEYRCDCAGCCDGWPCAMRPELLEKI-EALRCYFGRPVI 120
Query 57 ITSGYRTVSHNAKVGGARYSFHTRGMAADIRANGIKPKELAEVLNSI 103
ITSG R + N +VGG +SFHTRG AAD+ GI +LA+ +
Sbjct 121 ITSGVRCEARNEEVGGVSWSFHTRGCAADLYCPGIGVGDLAQTAKEL 167
> cjr:CJE0241 hypothetical protein
Length=129
Score = 66.2 bits (160), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 48/125 (38%), Positives = 62/125 (50%), Gaps = 13/125 (10%)
Query 15 KITSHFKVKEFACKDG----TPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKV 70
K +FK EF CK G V DE + ++ +I RE P+ I SGYR HNA+V
Sbjct 2 KNNPYFKESEFKCKCGKCELPQNVPSDELIDILCEI-REHYNAPVIINSGYRCKEHNAEV 60
Query 71 GGARYSFHTRGMAADIRANGIKPKELAE-VLNSIVPNSCGIIV-------YDNWVHFDTR 122
GGA S H G AAD G+K +E+ + VL + GI + Y +VH DTR
Sbjct 61 GGAPKSQHAIGSAADFVVKGVKTEEVHQYVLITYGERGLGIAIKHNFNDPYAGFVHLDTR 120
Query 123 NTKYR 127
K R
Sbjct 121 GKKAR 125
> cjs:CJS3_0707 hypothetical protein
Length=129
Score = 66.2 bits (160), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 48/125 (38%), Positives = 62/125 (50%), Gaps = 13/125 (10%)
Query 15 KITSHFKVKEFACKDG----TPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKV 70
K +FK EF CK G V DE + ++ +I RE P+ I SGYR HNA+V
Sbjct 2 KNNPYFKESEFKCKCGKCELPQNVPSDELIDILCEI-REHYNAPVIINSGYRCKEHNAEV 60
Query 71 GGARYSFHTRGMAADIRANGIKPKELAE-VLNSIVPNSCGIIV-------YDNWVHFDTR 122
GGA S H G AAD G+K +E+ + VL + GI + Y +VH DTR
Sbjct 61 GGAPKSQHAIGSAADFVVKGVKTEEVHQYVLITYGERGLGIAIKHNFNDPYAGFVHLDTR 120
Query 123 NTKYR 127
K R
Sbjct 121 GKKAR 125
> pput:L483_07785 peptidase M15
Length=143
Score = 65.9 bits (159), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 45/126 (36%), Positives = 62/126 (49%), Gaps = 13/126 (10%)
Query 16 ITSHFK-----VKEFACKDG---TPIVFVD---EYLAVILDIAREKIGKPITITSGYRTV 64
IT HF V + A +DG TP + L L+ R G PI ++SG+R+
Sbjct 3 ITPHFTLDEMTVSQLAARDGFDNTPPPDARANLQLLCCALEQVRALFGVPIIVSSGFRSE 62
Query 65 SHNAKVGGARYSFHTRGMAADIRANGIKPKELAEVLN-SIVPNSCGIIVYDNWVHFD-TR 122
N +GGA S H +G+AAD + P+E A +N S VP I+ +D WVH TR
Sbjct 63 KVNRLIGGATNSQHIQGLAADFTVMEVSPRETARRINESAVPFDQLILEFDRWVHLSVTR 122
Query 123 NTKYRK 128
T R+
Sbjct 123 GTPRRQ 128
> sba:Sulba_2498 Peptidase M15
Length=123
Score = 65.5 bits (158), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 36/108 (33%), Positives = 58/108 (54%), Gaps = 5/108 (5%)
Query 15 KITSHFKVKEFACKDGTPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVGGAR 74
K HFK +EF CK G + + L ++L+ AR+ P I S R +HN K+GG +
Sbjct 5 KTIKHFKKEEFTCKCGCSLNNISYTLVLMLEDARKLANMPFKINSACRCEAHNKKIGGVK 64
Query 75 YSFHTRGMAADIR--ANGIKPKELAEVLNSIVPNSCGIIVYDNWVHFD 120
S H +G+A DI ++G + L+ +LN C +++Y ++H D
Sbjct 65 DSAHVKGLAVDISIPSDGARFVILSALLNV---GFCRVLIYPTFIHVD 109
> gap:GAPWK_1388 hypothetical protein
Length=146
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 41/128 (32%), Positives = 65/128 (51%), Gaps = 12/128 (9%)
Query 15 KITSHFKVKEFACKDGTPIVFVDEYL-----------AVILDIAREKIGKPITITSGYRT 63
K+T HF ++EF + +D ++ A L+ RE + PI I+SGYR
Sbjct 2 KLTEHFTLEEFTRSTTATRLKIDNHVPDDLMVNIHLTANKLESVREALTNPIIISSGYRC 61
Query 64 VSHNAKVGGARYSFHTRGMAADIRANGIKPKELAE-VLNSIVPNSCGIIVYDNWVHFDTR 122
+ N KVGG++ S HT+G+A D PK++ + ++ + V I Y+ WVH +
Sbjct 62 PALNNKVGGSQNSAHTKGLAVDFHCAYGNPKQICQRLIMAGVEFDKIIQEYNQWVHIEFS 121
Query 123 NTKYRKGV 130
+T RK V
Sbjct 122 HTNQRKRV 129
> elm:ELI_1994 peptidase M15A
Length=190
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 35/86 (41%), Positives = 47/86 (55%), Gaps = 6/86 (7%)
Query 17 TSHFKVKEFACK-----DGTPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVG 71
T HF ++E+ C DG P V E L I + R G+P+ ITSG R + N +VG
Sbjct 83 TPHFAMEEYQCDCAGYCDGWPCVVEPELLEKI-EALRCYFGRPVIITSGVRCEARNEEVG 141
Query 72 GARYSFHTRGMAADIRANGIKPKELA 97
G +SFH RG AAD+ G+ +LA
Sbjct 142 GVSWSFHKRGCAADLYCPGVGVGDLA 167
> elm:ELI_3428 peptidase M15A
Length=186
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 34/86 (40%), Positives = 48/86 (56%), Gaps = 6/86 (7%)
Query 17 TSHFKVKEFACK-----DGTPIVFVDEYLAVILDIAREKIGKPITITSGYRTVSHNAKVG 71
++HF + E+AC DG P E L I + R G+P+ ITSG R + N +VG
Sbjct 76 SAHFLMDEYACDCAGDCDGWPAEMDPELLERI-EALRMACGQPVIITSGVRCAARNEEVG 134
Query 72 GARYSFHTRGMAADIRANGIKPKELA 97
G +SFH +GMAAD+ G+ +LA
Sbjct 135 GVAWSFHKKGMAADLYCPGMGIGDLA 160
> ppt:PPS_1672 peptidase M15A
Length=143
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 37/128 (29%), Positives = 59/128 (46%), Gaps = 12/128 (9%)
Query 15 KITSHFKVKEFACK--------DGTPIVFVDEYLAVI---LDIAREKIGKPITITSGYRT 63
+IT HF + E D P L ++ L+ R G P+ ++SGYR+
Sbjct 2 RITPHFTLDEMTVSQLAAREGLDNNPPAEARANLQLLCNALEQVRALFGAPVIVSSGYRS 61
Query 64 VSHNAKVGGARYSFHTRGMAADIRANGIKPKELA-EVLNSIVPNSCGIIVYDNWVHFDTR 122
+ N ++GG S H +G+AAD + P+E+ V S +P I+ +D+WVH
Sbjct 62 PAVNQRIGGTLTSKHLQGLAADFTVIDVSPREVVRRVSESTIPFDQLILEFDDWVHLAVS 121
Query 123 NTKYRKGV 130
R+ V
Sbjct 122 RGAPRRQV 129
> ble:BleG1_3098 Zinc D-Ala-D-Ala carboxypeptidase
Length=209
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 43/123 (35%), Positives = 60/123 (49%), Gaps = 14/123 (11%)
Query 11 QANEKITSHFKVKEFACKDGT-------PIVFVDE---YLAVILDIAREKIGK-PITITS 59
+A + T HF EF +DG+ V E L L+ R K+G P+TI S
Sbjct 79 EAADGSTRHFNWSEFHSRDGSRFSGGAVSATQVQENVRRLMYKLEAKRRKLGNIPLTINS 138
Query 60 GYRTVSHNAKVGGARYSFHTRGMAADIRANGIKPKELAEVLNSIVPNSCGIIVYDNWVHF 119
G+R+ SHN+ VGGA S H G AAD+ ++ P A + GII+Y + H
Sbjct 139 GFRSRSHNSSVGGASNSQHLYGSAADV-SSSTTPTRKANTARTC--GFSGIIIYQTFTHV 195
Query 120 DTR 122
D+R
Sbjct 196 DSR 198
> pmot:X970_06390 peptidase M15
Length=143
Score = 61.2 bits (147), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 36/128 (28%), Positives = 59/128 (46%), Gaps = 12/128 (9%)
Query 15 KITSHFKVKEFACK--------DGTPIVFVDEYLAVI---LDIAREKIGKPITITSGYRT 63
+IT HF + E D P L ++ L+ R G P+ ++SGYR+
Sbjct 2 RITPHFTLDEMTVSQLAAREGLDNNPPAEARANLQLLCNALEQVRALFGAPVIVSSGYRS 61
Query 64 VSHNAKVGGARYSFHTRGMAADIRANGIKPKELA-EVLNSIVPNSCGIIVYDNWVHFDTR 122
+ N ++GG S H +G+AAD + P+E+ + S +P I+ +D+WVH
Sbjct 62 PAVNQRIGGTLTSKHLQGLAADFTVIEVTPREVVRRISESTIPFDQLILEFDDWVHLAVS 121
Query 123 NTKYRKGV 130
R+ V
Sbjct 122 RGAPRRQV 129
Lambda K H a alpha
0.320 0.136 0.398 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 127666179182