bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-34_CDS_annotation_glimmer3.pl_2_4
Length=371
Score E
Sequences producing significant alignments: (Bits) Value
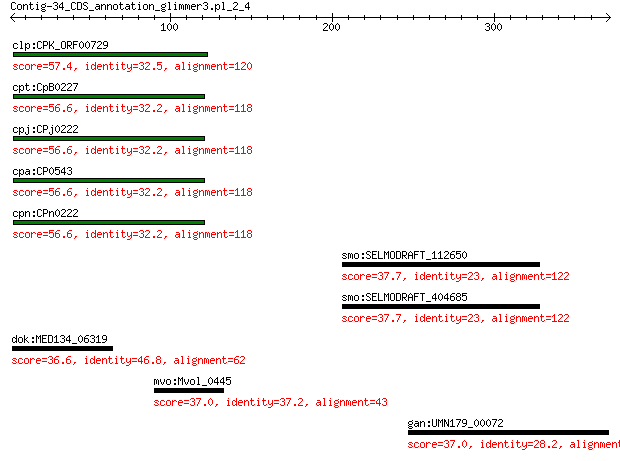
clp:CPK_ORF00729 hypothetical protein 57.4 2e-07
cpt:CpB0227 hypothetical protein 56.6 4e-07
cpj:CPj0222 hypothetical protein 56.6 4e-07
cpa:CP0543 hypothetical protein 56.6 4e-07
cpn:CPn0222 hypothetical protein 56.6 4e-07
smo:SELMODRAFT_112650 hypothetical protein 37.7 3.5
smo:SELMODRAFT_404685 hypothetical protein 37.7 3.6
dok:MED134_06319 hypothetical protein 36.6 6.7
mvo:Mvol_0445 hypothetical protein 37.0 7.3
gan:UMN179_00072 sn-glycerol-3-phosphate dehydrogenase subunit C 37.0 8.7
> clp:CPK_ORF00729 hypothetical protein
Length=121
Score = 57.4 bits (137), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 39/120 (33%), Positives = 54/120 (45%), Gaps = 35/120 (29%)
Query 3 CRLDRSRVWADRMLLELKDNDYKALFVTLTYNDRSLPSAWHVGSNYFDGFYEDVVPLVLD 62
CR ++VW+ R + E + K F+TLTY+D+ LP Y +V L L
Sbjct 30 CRTQHAKVWSYRCVHEASLYE-KNCFLTLTYDDKHLPQ------------YGSLVKLHL- 75
Query 63 DDEEWIAAAAGAPATLSIRDTQLFMKRLRKTFRDRRLRFFLAGEYGPKTHRPHYHAIIYG 122
QLF+KRLR ++R+F GEYG K RPHYH +I+
Sbjct 76 ---------------------QLFLKRLRDRISPHKIRYFGCGEYGTKLQRPHYHLLIFN 114
> cpt:CpB0227 hypothetical protein
Length=113
Score = 56.6 bits (135), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 38/118 (32%), Positives = 53/118 (45%), Gaps = 35/118 (30%)
Query 3 CRLDRSRVWADRMLLELKDNDYKALFVTLTYNDRSLPSAWHVGSNYFDGFYEDVVPLVLD 62
CR ++VW+ R + E + K F+TLTY+D+ LP Y +V L L
Sbjct 30 CRTQHAKVWSYRCVHEASLYE-KNCFLTLTYDDKHLPQ------------YGSLVKLHL- 75
Query 63 DDEEWIAAAAGAPATLSIRDTQLFMKRLRKTFRDRRLRFFLAGEYGPKTHRPHYHAII 120
QLF+KRLRK ++R+F G YG K RPHYH ++
Sbjct 76 ---------------------QLFLKRLRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpj:CPj0222 hypothetical protein
Length=113
Score = 56.6 bits (135), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 38/118 (32%), Positives = 53/118 (45%), Gaps = 35/118 (30%)
Query 3 CRLDRSRVWADRMLLELKDNDYKALFVTLTYNDRSLPSAWHVGSNYFDGFYEDVVPLVLD 62
CR ++VW+ R + E + K F+TLTY+D+ LP Y +V L L
Sbjct 30 CRTQHAKVWSYRCVHEASLYE-KNCFLTLTYDDKHLPQ------------YGSLVKLHL- 75
Query 63 DDEEWIAAAAGAPATLSIRDTQLFMKRLRKTFRDRRLRFFLAGEYGPKTHRPHYHAII 120
QLF+KRLRK ++R+F G YG K RPHYH ++
Sbjct 76 ---------------------QLFLKRLRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpa:CP0543 hypothetical protein
Length=113
Score = 56.6 bits (135), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 38/118 (32%), Positives = 53/118 (45%), Gaps = 35/118 (30%)
Query 3 CRLDRSRVWADRMLLELKDNDYKALFVTLTYNDRSLPSAWHVGSNYFDGFYEDVVPLVLD 62
CR ++VW+ R + E + K F+TLTY+D+ LP Y +V L L
Sbjct 30 CRTQHAKVWSYRCVHEASLYE-KNCFLTLTYDDKHLPQ------------YGSLVKLHL- 75
Query 63 DDEEWIAAAAGAPATLSIRDTQLFMKRLRKTFRDRRLRFFLAGEYGPKTHRPHYHAII 120
QLF+KRLRK ++R+F G YG K RPHYH ++
Sbjct 76 ---------------------QLFLKRLRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpn:CPn0222 hypothetical protein
Length=113
Score = 56.6 bits (135), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 38/118 (32%), Positives = 53/118 (45%), Gaps = 35/118 (30%)
Query 3 CRLDRSRVWADRMLLELKDNDYKALFVTLTYNDRSLPSAWHVGSNYFDGFYEDVVPLVLD 62
CR ++VW+ R + E + K F+TLTY+D+ LP Y +V L L
Sbjct 30 CRTQHAKVWSYRCVHEASLYE-KNCFLTLTYDDKHLPQ------------YGSLVKLHL- 75
Query 63 DDEEWIAAAAGAPATLSIRDTQLFMKRLRKTFRDRRLRFFLAGEYGPKTHRPHYHAII 120
QLF+KRLRK ++R+F G YG K RPHYH ++
Sbjct 76 ---------------------QLFLKRLRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> smo:SELMODRAFT_112650 hypothetical protein
Length=509
Score = 37.7 bits (86), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 28/123 (23%), Positives = 54/123 (44%), Gaps = 1/123 (1%)
Query 206 GLLHADDLLKKGDKTFIRDIDLNGKECTREVYLGRAFIRSAAREHMKPVFAAADLVESVQ 265
GL++ D++L++ D + DL + T +++L + + KPV A ++ES+
Sbjct 246 GLVNFDEILRESDAIMVARGDLGMEIPTEKIFLAQKMMIYKCNAAGKPVITATQMLESMI 305
Query 266 TCINELEAESCTEHEDVLDALYICLRAFRHRLDDVIELHIPPSVRLCSR-QNERAYNSIQ 324
C AE+ VLD + + EL + ++C +N Y +I
Sbjct 306 KCPRPTRAEATDVANAVLDGTDAVMLSGETAAGLYPELAVATMAKICVEAENSLDYPAIF 365
Query 325 RSI 327
++I
Sbjct 366 KAI 368
> smo:SELMODRAFT_404685 hypothetical protein
Length=509
Score = 37.7 bits (86), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 28/123 (23%), Positives = 54/123 (44%), Gaps = 1/123 (1%)
Query 206 GLLHADDLLKKGDKTFIRDIDLNGKECTREVYLGRAFIRSAAREHMKPVFAAADLVESVQ 265
GL++ D++L++ D + DL + T +++L + + KPV A ++ES+
Sbjct 246 GLVNFDEILRESDAIMVARGDLGMEIPTEKIFLAQKMMIYKCNAAGKPVITATQMLESMI 305
Query 266 TCINELEAESCTEHEDVLDALYICLRAFRHRLDDVIELHIPPSVRLCSR-QNERAYNSIQ 324
C AE+ VLD + + EL + ++C +N Y +I
Sbjct 306 KCPRPTRAEATDVANAVLDGTDAVMLSGETAAGLYPELAVATMAKICVEAENSLDYPAIF 365
Query 325 RSI 327
++I
Sbjct 366 KAI 368
> dok:MED134_06319 hypothetical protein
Length=288
Score = 36.6 bits (83), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 29/78 (37%), Positives = 38/78 (49%), Gaps = 16/78 (21%)
Query 2 GCRLDRSRVWADRMLLE---------LKDNDYKALFVTLTYNDRSLPSAWHVGSN--YFD 50
GC +R RV + E LK++ Y AL+ T T +RS+ S GSN YFD
Sbjct 68 GCGTERPRVLMRSVFDEIFTQATDIVLKEDTYAALYATATEGERSIVSILGTGSNCSYFD 127
Query 51 G--FYEDVVPL---VLDD 63
G + VV L V+DD
Sbjct 128 GTDIIQKVVSLGYVVMDD 145
> mvo:Mvol_0445 hypothetical protein
Length=493
Score = 37.0 bits (84), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 25/43 (58%), Gaps = 2/43 (5%)
Query 90 LRKTFRDRRLRFFLAGEYGPKTHRPHYHAIIYGLTLSDFKDCR 132
+RKT +R+L + + E+ PK PH H +I+G D +D R
Sbjct 256 IRKT--NRKLEYIYSFEFSPKKALPHLHVVIFGTDFLDLRDYR 296
> gan:UMN179_00072 sn-glycerol-3-phosphate dehydrogenase subunit
C
Length=1022
Score = 37.0 bits (84), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 35/139 (25%), Positives = 58/139 (42%), Gaps = 23/139 (17%)
Query 247 AREHMKPVFAAADLVESVQTCIN-ELEAESCTEHEDVLDALYICLRAFRHRLDDVIELHI 305
A+ H KP+ D TC+ E A+ TE +LD ++ F H D LH+
Sbjct 416 AKGHAKPIPFVED------TCVPPENLADYITEFRALLDKHHLEYGMFGHV--DAGVLHV 467
Query 306 PPSVRLCSRQNERAYNSIQRSISNL--------------AVFGGYLLEYYQGKELHFLQR 351
P++ LC + + + SI +++L V G Y +++ + LQ+
Sbjct 468 RPALDLCDSEQVKLFKSISDQVADLTQKYGGLIWGEHGKGVRGEYSRKFFGEQLWQVLQQ 527
Query 352 IKLLPERGDLFEQTSTCKP 370
IK L + + CKP
Sbjct 528 IKTLFDPDNRLNPGKICKP 546
Lambda K H a alpha
0.324 0.139 0.435 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 716235340540