bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-32_CDS_annotation_glimmer3.pl_2_5
Length=125
Score E
Sequences producing significant alignments: (Bits) Value
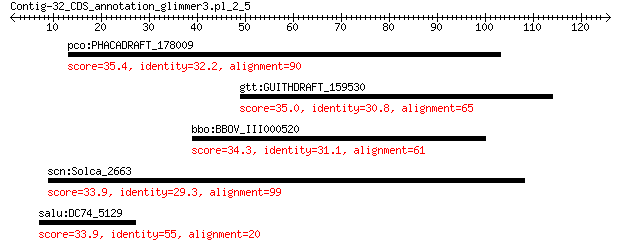
pco:PHACADRAFT_178009 hypothetical protein 35.4 2.1
gtt:GUITHDRAFT_159530 hypothetical protein 35.0 2.9
bbo:BBOV_III000520 17.m07071; PHD-finger family protein 34.3 5.6
scn:Solca_2663 Bacteroidetes-specific putative membrane protein 33.9 6.0
salu:DC74_5129 hypothetical protein 33.9 6.5
> pco:PHACADRAFT_178009 hypothetical protein
Length=381
Score = 35.4 bits (80), Expect = 2.1, Method: Composition-based stats.
Identities = 29/90 (32%), Positives = 43/90 (48%), Gaps = 10/90 (11%)
Query 13 ESLRFYTDEDGCVHMNSDVNLLMNAERIRNQIGEENYLNIVRSIQPSRSPYKQKLDDDAL 72
ES R +E G + D L + RI +I E N+L ++S PS +P+ DDA
Sbjct 108 ESRRQLKEELGAKTLPPDHPLTRHVHRIVTRILEANHLGHLKSSTPSTAPWSAGTHDDA- 166
Query 73 ISTLKSRYLQSPAEIDAWMASLDQKYEDMV 102
YL A++ A S D+++E MV
Sbjct 167 -------YLTPEAQLPA--GSGDKEWELMV 187
> gtt:GUITHDRAFT_159530 hypothetical protein
Length=4460
Score = 35.0 bits (79), Expect = 2.9, Method: Composition-based stats.
Identities = 20/65 (31%), Positives = 35/65 (54%), Gaps = 0/65 (0%)
Query 49 YLNIVRSIQPSRSPYKQKLDDDALISTLKSRYLQSPAEIDAWMASLDQKYEDMVAEVKAA 108
Y +IV +++P R KQ D+ +T + A+++A +A L Q+Y+ +VAE A
Sbjct 3162 YYDIVVTVEPKRQALKQAQDELEAANTKLAEVNAHVADLEAKLAVLIQEYDKVVAEKNAV 3221
Query 109 AAASQ 113
A +
Sbjct 3222 VAEGE 3226
> bbo:BBOV_III000520 17.m07071; PHD-finger family protein
Length=2204
Score = 34.3 bits (77), Expect = 5.6, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 34/61 (56%), Gaps = 2/61 (3%)
Query 39 RIRNQIGEENYLNIVRSIQPSRSPYKQKLDDDALISTLKSRYLQSPAEIDAWMASLDQKY 98
+ R+ +G ++YLN +I S P K+ DDD I+ + Y+ + EID +S+ ++
Sbjct 383 KARSDMGMQSYLNPYNAIADSLVPLKRSTDDD--ITAVIREYVTTLEEIDVAFSSIIERV 440
Query 99 E 99
E
Sbjct 441 E 441
> scn:Solca_2663 Bacteroidetes-specific putative membrane protein
Length=303
Score = 33.9 bits (76), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 29/102 (28%), Positives = 47/102 (46%), Gaps = 5/102 (5%)
Query 9 IQPIESLRFYTDED-GCVHMNSDVNLLMNAERIRNQIGEENYLNIVRSIQPSRSPYKQKL 67
+Q S Y D D G + S + L IRN I +E Y I RS+ S + YK +
Sbjct 150 LQEYNSRGAYMDGDFGIAYTGSGLTLQAAMPTIRNYIVKETYSTIDRSVLYSAASYKINM 209
Query 68 DDDALISTL--KSRYLQSPAEIDAWMASLDQKYEDMVAEVKA 107
++ +S++ K Y D W A ++ K +A+++A
Sbjct 210 GEE--LSSIEPKLSYTMVKGNTDIWDAGVNVKVASNLADIQA 249
> salu:DC74_5129 hypothetical protein
Length=554
Score = 33.9 bits (76), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 11/20 (55%), Positives = 17/20 (85%), Gaps = 0/20 (0%)
Query 7 VVIQPIESLRFYTDEDGCVH 26
+V++ +ES RFY D++GCVH
Sbjct 98 MVVKELESERFYVDDEGCVH 117
Lambda K H a alpha
0.311 0.125 0.339 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 125791032939