bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_5
Length=217
Score E
Sequences producing significant alignments: (Bits) Value
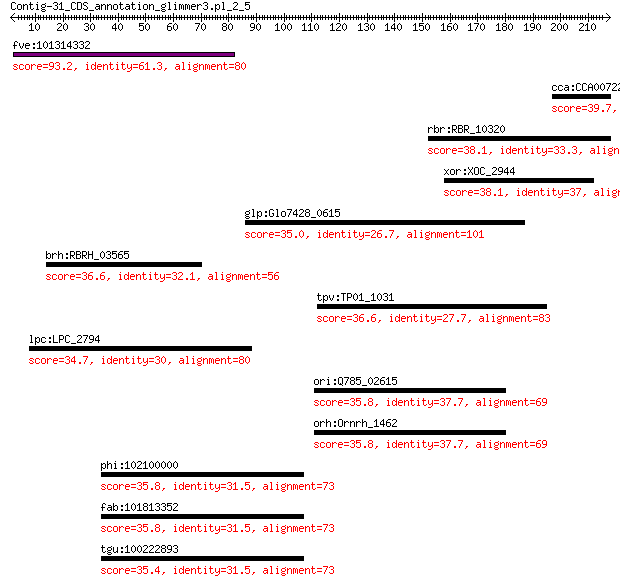
fve:101314332 capsid protein VP1-like 93.2 2e-19
cca:CCA00722 hypothetical protein 39.7 0.072
rbr:RBR_10320 Signal transduction histidine kinase 38.1 1.1
xor:XOC_2944 putative signal protein with HAMP, GGDEF and EAL ... 38.1 1.3
glp:Glo7428_0615 Endoribonuclease L-PSP 35.0 3.8
brh:RBRH_03565 UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyl... 36.6 3.8
tpv:TP01_1031 hypothetical protein 36.6 3.8
lpc:LPC_2794 traJ; conjugal transfer relaxosome component TraJ 34.7 4.8
ori:Q785_02615 hypothetical protein 35.8 6.7
orh:Ornrh_1462 hypothetical protein 35.8 6.7
phi:102100000 EXD2; exonuclease 3'-5' domain containing 2 35.8
fab:101813352 EXD2; exonuclease 3'-5' domain containing 2 35.8
tgu:100222893 EXD2; exonuclease 3'-5' domain containing 2 35.4
> fve:101314332 capsid protein VP1-like
Length=421
Score = 93.2 bits (230), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 49/83 (59%), Positives = 59/83 (71%), Gaps = 3/83 (4%)
Query 2 FNVISPDARLQRPEYLGGTHSRVNVVPTAQTSSTDSVS---PQSNLSAFGVLGDSAHGFN 58
F V SPDARLQRPEYLGG + +N+ P AQT T + PQ NL+AFG HGF+
Sbjct 339 FGVASPDARLQRPEYLGGGSTPINIAPIAQTGGTGAQGTTTPQGNLAAFGTYMAKGHGFS 398
Query 59 KSFVEHGYVIGLCCLRADITYQQ 81
+SFVEHG+VIGL +RAD+TYQQ
Sbjct 399 QSFVEHGHVIGLVSVRADLTYQQ 421
> cca:CCA00722 hypothetical protein
Length=117
Score = 39.7 bits (91), Expect = 0.072, Method: Compositional matrix adjust.
Identities = 15/21 (71%), Positives = 18/21 (86%), Gaps = 0/21 (0%)
Query 197 FDLKTSRPMPVYSVPGLVDHF 217
F L+ +RPMPVYSV GL+DHF
Sbjct 97 FSLRCARPMPVYSVSGLIDHF 117
> rbr:RBR_10320 Signal transduction histidine kinase
Length=479
Score = 38.1 bits (87), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 34/66 (52%), Gaps = 9/66 (14%)
Query 152 SLDVWHLAQRFDSLPKLNQDFIEENPPINRVIAVQNEPQFFADFWFDLKTSRPMPVYSVP 211
S ++ L+Q F+++ +D+IE+ +A QN F ADF +LKT P+ SV
Sbjct 216 STEITELSQSFNTMADYVEDYIEQLK-----LATQNRDNFIADFTHELKT----PLTSVI 266
Query 212 GLVDHF 217
G D
Sbjct 267 GYADML 272
> xor:XOC_2944 putative signal protein with HAMP, GGDEF and EAL
domains
Length=785
Score = 38.1 bits (87), Expect = 1.3, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 29/54 (54%), Gaps = 1/54 (2%)
Query 158 LAQRFDSLPKLNQDFIEENPPINRVIAVQNEPQFFADFWFDLKTSRPMPVYSVP 211
LAQ F S L+ F + PP+ RV+A EP A+F L ++ +P S+P
Sbjct 176 LAQDFRSTTGLDATFFTDGPPL-RVLASTMEPSARAEFAQQLAAAKKLPAVSIP 228
> glp:Glo7428_0615 Endoribonuclease L-PSP
Length=131
Score = 35.0 bits (79), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 27/110 (25%), Positives = 46/110 (42%), Gaps = 9/110 (8%)
Query 86 MWSRRQLFDFYWPTLAHLGEQVVYNKEIYAQGTA---DDNGVFGYQERYAEYRYKPSMIT 142
M +R W T A + IY GTA + GVF + YA+ + ++I
Sbjct 1 MQRQRTFSGTAWETKAGYCRAIRVGNHIYVSGTAPVDEQGGVFAPNDAYAQTKRCLAIIQ 60
Query 143 GKLRSTDAQSLDVWHL------AQRFDSLPKLNQDFIEENPPINRVIAVQ 186
L++ A DV +R+ + +Q+ E+PP N ++ +Q
Sbjct 61 KALQNLGADLPDVVRTRVYVTDMKRWAEFAQAHQESFGEHPPANTMVEIQ 110
> brh:RBRH_03565 UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase
(EC:2.3.1.-)
Length=378
Score = 36.6 bits (83), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 28/56 (50%), Gaps = 0/56 (0%)
Query 14 PEYLGGTHSRVNVVPTAQTSSTDSVSPQSNLSAFGVLGDSAHGFNKSFVEHGYVIG 69
P L G H V P A+ +++ ++ P + A V+G+ +FV HG VIG
Sbjct 113 PTALAGIHPGAVVDPAAKVAASATIGPHVTIEAGAVIGERVRIDAHAFVGHGAVIG 168
> tpv:TP01_1031 hypothetical protein
Length=665
Score = 36.6 bits (83), Expect = 3.8, Method: Composition-based stats.
Identities = 23/84 (27%), Positives = 42/84 (50%), Gaps = 3/84 (4%)
Query 112 EIYAQGTADDNGVFGYQERYAEYRYKPSMITGKLRSTDAQSLDVWHLAQRFDSLPK-LNQ 170
++Y + D+ +F E Y +YK S++ G LR ++ + +L+ + S+ +N
Sbjct 310 KVYKRSQVDEGSLFELVEEYCNQKYKESIVMGHLRGKMSKIPE--NLSNLWASMTDCINS 367
Query 171 DFIEENPPINRVIAVQNEPQFFAD 194
+ I PIN+VI PQ +D
Sbjct 368 ELILRLRPINKVITNHPFPQTRSD 391
> lpc:LPC_2794 traJ; conjugal transfer relaxosome component TraJ
Length=117
Score = 34.7 bits (78), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 24/81 (30%), Positives = 39/81 (48%), Gaps = 2/81 (2%)
Query 8 DARLQRPEYLGGTHSRVNVVPTAQTSSTDSVSPQSNLSAFGVLGDSAHGFN-KSFVEHGY 66
D + + P G H RV V+P + S S + Q+ LS G L G+ S ++ Y
Sbjct 2 DDKNKSPSRKHGRHLRVPVLPDEEIS-IKSHAAQAGLSVAGYLRRIGLGYQIHSAIDKDY 60
Query 67 VIGLCCLRADITYQQGLNRMW 87
++ L + AD+ GL ++W
Sbjct 61 ILQLSKINADMGRLGGLFKLW 81
> ori:Q785_02615 hypothetical protein
Length=383
Score = 35.8 bits (81), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 26/79 (33%), Positives = 37/79 (47%), Gaps = 18/79 (23%)
Query 111 KEIYAQGTADDNGVFGYQERYAEYRYKPSMITGKLRSTDAQSL-DVWH---LAQRFDSL- 165
+EI+ + + G FGY Y P + GK R + SL V H + Q + L
Sbjct 310 QEIFQEAAQNHYGTFGY--------YSPMIFLGKKRYVEDSSLYSVLHQLAIGQEYKKLL 361
Query 166 -----PKLNQDFIEENPPI 179
P+L DFI++NPPI
Sbjct 362 YLTDEPELIVDFIKKNPPI 380
> orh:Ornrh_1462 hypothetical protein
Length=383
Score = 35.8 bits (81), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 26/79 (33%), Positives = 37/79 (47%), Gaps = 18/79 (23%)
Query 111 KEIYAQGTADDNGVFGYQERYAEYRYKPSMITGKLRSTDAQSL-DVWH---LAQRFDSL- 165
+EI+ + + G FGY Y P + GK R + SL V H + Q + L
Sbjct 310 QEIFQEAAQNHYGTFGY--------YSPMIFLGKKRYVEDSSLYSVLHQLAIGQEYKKLL 361
Query 166 -----PKLNQDFIEENPPI 179
P+L DFI++NPPI
Sbjct 362 YLTDEPELIVDFIKKNPPI 380
> phi:102100000 EXD2; exonuclease 3'-5' domain containing 2
Length=621
Score = 35.8 bits (81), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 23/73 (32%), Positives = 35/73 (48%), Gaps = 6/73 (8%)
Query 34 STDSVSPQSNLSAFGVLGDSAHGFNKSFVEHGYVIGLCCLRADITYQQGLNRMWSRRQLF 93
+TD+V+P+ +A G+ N+S+V HG + CC + + L R W R+
Sbjct 532 NTDTVTPEMLQAAAGL---ETRICNESYVPHGLKVVQCCAKGGLRSLMQLERRW-RQHFL 587
Query 94 DFYWPTLAHLGEQ 106
D P HL EQ
Sbjct 588 DSMKPK--HLPEQ 598
> fab:101813352 EXD2; exonuclease 3'-5' domain containing 2
Length=621
Score = 35.8 bits (81), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 23/73 (32%), Positives = 35/73 (48%), Gaps = 6/73 (8%)
Query 34 STDSVSPQSNLSAFGVLGDSAHGFNKSFVEHGYVIGLCCLRADITYQQGLNRMWSRRQLF 93
+TD+V+P+ +A G+ N+S+V HG + CC + + L R W R+
Sbjct 532 NTDTVTPEMLQTAAGL---ETRICNESYVPHGLKVVQCCAKGGLRSLMQLERRW-RQHFL 587
Query 94 DFYWPTLAHLGEQ 106
D P HL EQ
Sbjct 588 DSMKPK--HLPEQ 598
> tgu:100222893 EXD2; exonuclease 3'-5' domain containing 2
Length=621
Score = 35.4 bits (80), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 23/73 (32%), Positives = 35/73 (48%), Gaps = 6/73 (8%)
Query 34 STDSVSPQSNLSAFGVLGDSAHGFNKSFVEHGYVIGLCCLRADITYQQGLNRMWSRRQLF 93
+TD+V+P+ +A G+ N+S+V HG + CC + + L R W R+
Sbjct 532 NTDTVTPEMLQAAAGL---ETRICNESYVPHGLKVVQCCAKGGLRSLMQLERRW-RQHFL 587
Query 94 DFYWPTLAHLGEQ 106
D P HL EQ
Sbjct 588 DSMKPK--HLPEQ 598
Lambda K H a alpha
0.321 0.137 0.426 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 266741246099