bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_4
Length=259
Score E
Sequences producing significant alignments: (Bits) Value
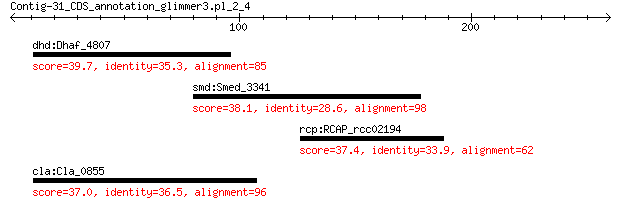
dhd:Dhaf_4807 phage tape measure protein 39.7 0.58
smd:Smed_3341 purine nucleoside phosphorylase 38.1 1.1
rcp:RCAP_rcc02194 phospholipase/carboxylesterase (EC:3.1.-.-) 37.4 1.8
cla:Cla_0855 hypothetical protein 37.0 4.1
> dhd:Dhaf_4807 phage tape measure protein
Length=694
Score = 39.7 bits (91), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 30/91 (33%), Positives = 42/91 (46%), Gaps = 16/91 (18%)
Query 11 GGAIGGLFGDAIGGTITGATLGSHLGS------IGSALGTGMGLVGSAKGLYDSFNNTSL 64
G G +F +A+ G I+GA +GS +G +G A G G+GL+ N T+
Sbjct 162 GSDAGTMFQNALSGVISGAAIGSAVGPPVIGTVVGIAAGLGLGLI----------NGTTS 211
Query 65 KNQMAYDQFKSELDYQYWSRKMSNRHTLEVG 95
N D FK + QY + K S TL G
Sbjct 212 INNKKDDAFKDYVQEQYNTIKQSQEDTLAHG 242
> smd:Smed_3341 purine nucleoside phosphorylase
Length=265
Score = 38.1 bits (87), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 50/103 (49%), Gaps = 12/103 (12%)
Query 80 QYWSRKMSNRHTLEVGDLRQAGLNPILSANSAGSVASAIPNGAIPETSSQQSAAGAAREA 139
Y+ R +N + + L++ G++ ++ NSAGS+ +P G++ + + AGA
Sbjct 78 HYYERGDANAMRVPIETLKRIGVDNLILTNSAGSLREDMPPGSVMRIADHIAFAGA---- 133
Query 140 NRINAMIGESTSAK-----NLADAQASIMNAETGRMVGIASAR 177
N +IG + A+ N DA ++ ET +GI AR
Sbjct 134 ---NPLIGVESDARFVGMTNAYDAALAVGMEETAERLGIPLAR 173
> rcp:RCAP_rcc02194 phospholipase/carboxylesterase (EC:3.1.-.-)
Length=220
Score = 37.4 bits (85), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 38/63 (60%), Gaps = 1/63 (2%)
Query 126 TSSQQSAAGAAREANRINAMIGESTSAKNLADAQASIMNAETGRMVGIASA-RRANAEAG 184
++ +++AAG R + +NA + + +A+ LA AQ +++ G M+ + A RRA A AG
Sbjct 77 SAPEEAAAGLTRSSEDLNAFLDQVMAAEGLAAAQIALLGFSQGTMMALQVAPRRAQALAG 136
Query 185 LAG 187
+ G
Sbjct 137 VVG 139
> cla:Cla_0855 hypothetical protein
Length=1829
Score = 37.0 bits (84), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 35/102 (34%), Positives = 45/102 (44%), Gaps = 14/102 (14%)
Query 11 GGAIGGLFGDAIG---GTITGATLGSHLGSI-GSALGTGMGLVGSAKGLYDSFNNTSLKN 66
GGAIGG+ G I GT+ G LGS +G I G TG GL L+ N K+
Sbjct 1090 GGAIGGVLGSLIAPGIGTLVGGLLGSVIGGIFGKTKATGSGL-----RLWQDIN---FKD 1141
Query 67 QMAYDQFKSELDYQY--WSRKMSNRHTLEVGDLRQAGLNPIL 106
+ +S +DYQ W K S + D + LN L
Sbjct 1142 HFSQGNLQSYIDYQKKGWFSKKSWTEYQNINDRKIKELNLFL 1183
Lambda K H a alpha
0.313 0.128 0.365 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 386865484076