bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_3
Length=315
Score E
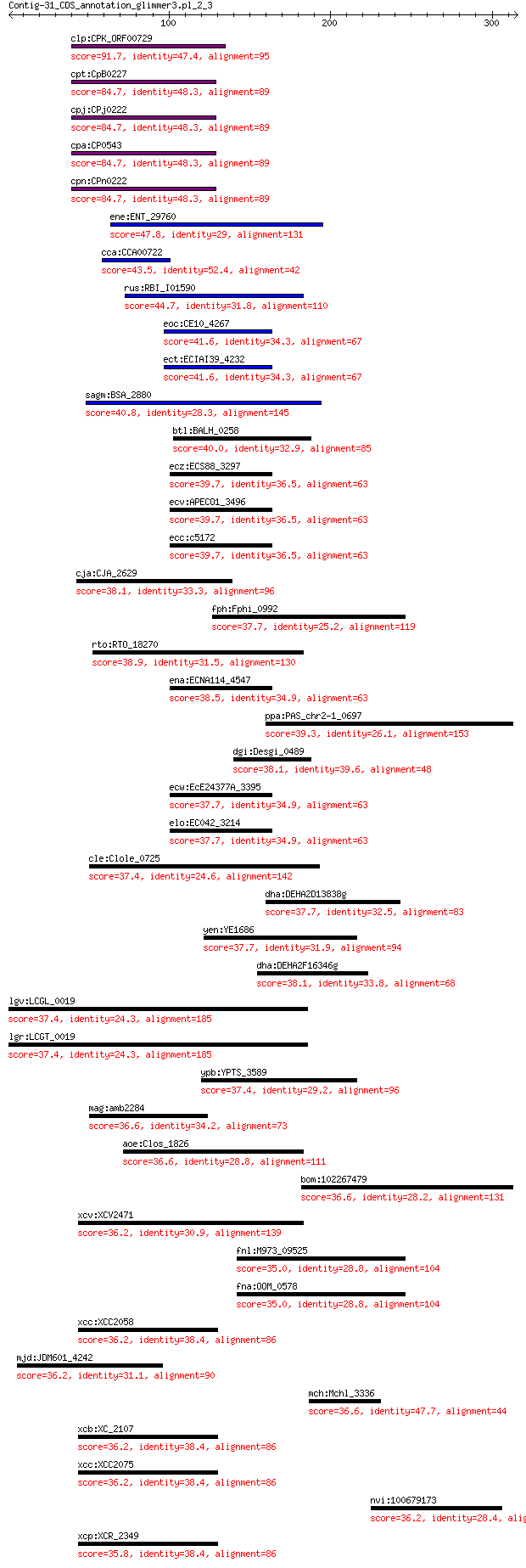
Sequences producing significant alignments: (Bits) Value
clp:CPK_ORF00729 hypothetical protein 91.7 7e-20
cpt:CpB0227 hypothetical protein 84.7 2e-17
cpj:CPj0222 hypothetical protein 84.7 2e-17
cpa:CP0543 hypothetical protein 84.7 2e-17
cpn:CPn0222 hypothetical protein 84.7 2e-17
ene:ENT_29760 hypothetical protein 47.8 0.001
cca:CCA00722 hypothetical protein 43.5 0.007
rus:RBI_I01590 hypothetical phage protein 44.7 0.014
eoc:CE10_4267 hypothetical protein 41.6 0.12
ect:ECIAI39_4232 hypothetical protein 41.6 0.12
sagm:BSA_2880 Rep protein 40.8 0.19
btl:BALH_0258 Rep protein 40.0 0.44
ecz:ECS88_3297 hypothetical protein 39.7 0.50
ecv:APECO1_3496 hypothetical protein 39.7 0.50
ecc:c5172 hypothetical protein 39.7 0.50
cja:CJA_2629 hypothetical protein 38.1 0.89
fph:Fphi_0992 hypothetical protein 37.7 0.89
rto:RTO_18270 hypothetical protein 38.9 0.95
ena:ECNA114_4547 hypothetical protein 38.5 1.0
ppa:PAS_chr2-1_0697 Catalytic subunit of DNA polymerase zeta, ... 39.3 1.2
dgi:Desgi_0489 glycyl-radical enzyme activator family protein 38.1 2.0
ecw:EcE24377A_3395 hypothetical protein 37.7 2.0
elo:EC042_3214 hypothetical protein 37.7 2.0
cle:Clole_0725 hypothetical protein 37.4 2.7
dha:DEHA2D13838g DEHA2D13838p 37.7 2.7
yen:YE1686 gpA; phage replication protein 37.7 2.9
dha:DEHA2F16346g DEHA2F16346p 38.1 3.0
lgv:LCGL_0019 hypothetical protein 37.4 3.0
lgr:LCGT_0019 hypothetical protein 37.4 3.0
ypb:YPTS_3589 replication gene A 37.4 4.4
mag:amb2284 response regulator containing CheY-like receiver 36.6 6.1
aoe:Clos_1826 hypothetical protein 36.6 6.3
bom:102267479 FSTL4; follistatin-like 4 36.6
xcv:XCV2471 filamentous phage phiLf replication initiation pro... 36.2 7.7
fnl:M973_09525 peptidase 35.0 8.1
fna:OOM_0578 hypothetical protein 35.0 8.1
xcc:XCC2058 gII; replication initiation protein 36.2 8.7
mjd:JDM601_4242 tesB2; acyl-CoA thioesterase 36.2 8.9
mch:Mchl_3336 PAS/PAC sensor hybrid histidine kinase 36.6 9.0
xcb:XC_2107 replication initiation protein 36.2 9.2
xcc:XCC2075 gII; replication initiation protein 36.2 9.2
nvi:100679173 G-protein coupled receptor Mth2-like 36.2 9.4
xcp:XCR_2349 filamentous phage phiLf replication initiation pr... 35.8 10.0
> clp:CPK_ORF00729 hypothetical protein
Length=121
Score = 91.7 bits (226), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 45/97 (46%), Positives = 61/97 (63%), Gaps = 2/97 (2%)
Query 40 IPCGQCIGCRLDRSLDSAVRAHHESLLYDRNYFLTLTYSPEHLPPFGSLIPRDLTLFWKR 99
+PC +C CR + + R HE+ LY++N FLTLTY +HLP +GSL+ L LF KR
Sbjct 22 MPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDKHLPQYGSLVKLHLQLFLKR 81
Query 100 LRKRGV--SLRYMACGEYGSAFGRPHYHAILFNLPAI 134
LR R +RY CGEYG+ RPHYH ++FN ++
Sbjct 82 LRDRISPHKIRYFGCGEYGTKLQRPHYHLLIFNYDSL 118
> cpt:CpB0227 hypothetical protein
Length=113
Score = 84.7 bits (208), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 43/91 (47%), Positives = 56/91 (62%), Gaps = 2/91 (2%)
Query 40 IPCGQCIGCRLDRSLDSAVRAHHESLLYDRNYFLTLTYSPEHLPPFGSLIPRDLTLFWKR 99
+PC +C CR + + R HE+ LY++N FLTLTY +HLP +GSL+ L LF KR
Sbjct 22 MPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDKHLPQYGSLVKLHLQLFLKR 81
Query 100 LRKRGV--SLRYMACGEYGSAFGRPHYHAIL 128
LRK +RY CG YG+ RPHYH +L
Sbjct 82 LRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpj:CPj0222 hypothetical protein
Length=113
Score = 84.7 bits (208), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 43/91 (47%), Positives = 56/91 (62%), Gaps = 2/91 (2%)
Query 40 IPCGQCIGCRLDRSLDSAVRAHHESLLYDRNYFLTLTYSPEHLPPFGSLIPRDLTLFWKR 99
+PC +C CR + + R HE+ LY++N FLTLTY +HLP +GSL+ L LF KR
Sbjct 22 MPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDKHLPQYGSLVKLHLQLFLKR 81
Query 100 LRKRGV--SLRYMACGEYGSAFGRPHYHAIL 128
LRK +RY CG YG+ RPHYH +L
Sbjct 82 LRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpa:CP0543 hypothetical protein
Length=113
Score = 84.7 bits (208), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 43/91 (47%), Positives = 56/91 (62%), Gaps = 2/91 (2%)
Query 40 IPCGQCIGCRLDRSLDSAVRAHHESLLYDRNYFLTLTYSPEHLPPFGSLIPRDLTLFWKR 99
+PC +C CR + + R HE+ LY++N FLTLTY +HLP +GSL+ L LF KR
Sbjct 22 MPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDKHLPQYGSLVKLHLQLFLKR 81
Query 100 LRKRGV--SLRYMACGEYGSAFGRPHYHAIL 128
LRK +RY CG YG+ RPHYH +L
Sbjct 82 LRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpn:CPn0222 hypothetical protein
Length=113
Score = 84.7 bits (208), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 43/91 (47%), Positives = 56/91 (62%), Gaps = 2/91 (2%)
Query 40 IPCGQCIGCRLDRSLDSAVRAHHESLLYDRNYFLTLTYSPEHLPPFGSLIPRDLTLFWKR 99
+PC +C CR + + R HE+ LY++N FLTLTY +HLP +GSL+ L LF KR
Sbjct 22 MPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDKHLPQYGSLVKLHLQLFLKR 81
Query 100 LRKRGV--SLRYMACGEYGSAFGRPHYHAIL 128
LRK +RY CG YG+ RPHYH +L
Sbjct 82 LRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> ene:ENT_29760 hypothetical protein
Length=299
Score = 47.8 bits (112), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 38/138 (28%), Positives = 69/138 (50%), Gaps = 11/138 (8%)
Query 64 SLLYDRNYFLTLTYSPEHLPPFGSLIPRDLTLFW-KRLRKRGVSLRYMACGEYGSAFGRP 122
+L D +F TLT P+ ++L L W +++R R + Y+ E + G
Sbjct 80 ALANDFKWFTTLTIDPKQYDSLDYDSSKELLLKWCRKMRDRYENFDYLIVPELHKS-GAV 138
Query 123 HYHAILFNLPA--IELK--QIGTTHTRFPTYISNVISECWPFGFHTINSVSFE--TCAYV 176
H+H++L ++PA IE K + G R I N+ W +GF + + T +Y+
Sbjct 139 HFHSLLGDIPANFIEAKHPKTGAPLLRNERQIYNLAD--WTYGFTDCEEIEDKERTASYL 196
Query 177 ARYVTKKILGDGKQVYEK 194
+Y+TK+++ D K+++ K
Sbjct 197 TKYITKELMTD-KEMFRK 213
> cca:CCA00722 hypothetical protein
Length=117
Score = 43.5 bits (101), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 22/42 (52%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 59 RAHHESLLYDRNYFLTLTYSPEHLPPFGSLIPRDLTLFWKRL 100
R HE+ LY +N FLTLTY +LP GSL+ RD+ LF R
Sbjct 49 RCIHEASLYGQNSFLTLTYEDRNLPEKGSLVRRDVRLFLMRF 90
> rus:RBI_I01590 hypothetical phage protein
Length=271
Score = 44.7 bits (104), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 35/119 (29%), Positives = 54/119 (45%), Gaps = 27/119 (23%)
Query 73 LTLTYSPEHLPPFGSLIPRDLTLFWKRLR----KRGV-SLRYMACGEYGSAFGRPHYHAI 127
+ LTY+ +HLP + + F RL+ K G+ SL+YM+ E GS GR H+H I
Sbjct 69 IHLTYNGDHLPGNDEAVKKQFRNFIARLKRYRKKHGLPSLKYMSVTERGSRNGRYHHHTI 128
Query 128 L--FNLPAIELKQIGTTHTRFPTYISNVISECWPFGFHTINSVSFETCAY--VARYVTK 182
+ ++PA V+ E W G+ I + F+ C ++RY K
Sbjct 129 VNCGDMPAP------------------VLVELWGQGYVDIKVLQFDQCGVEGLSRYFCK 169
> eoc:CE10_4267 hypothetical protein
Length=243
Score = 41.6 bits (96), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 30/67 (45%), Gaps = 3/67 (4%)
Query 97 WKRLRKRGVSLRYMACGEYGSAFGRPHYHAILFNLPAIELKQIGTTHTRFPTYISNVISE 156
W R SLRY C E+G +GR HYH IL L +G P+ ++ +I E
Sbjct 75 WANQRVHATSLRYFWCREFGKMYGRKHYHVILL-LNKDTWCSLGDFSE--PSSLATMIQE 131
Query 157 CWPFGFH 163
W H
Sbjct 132 AWCSALH 138
> ect:ECIAI39_4232 hypothetical protein
Length=243
Score = 41.6 bits (96), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 30/67 (45%), Gaps = 3/67 (4%)
Query 97 WKRLRKRGVSLRYMACGEYGSAFGRPHYHAILFNLPAIELKQIGTTHTRFPTYISNVISE 156
W R SLRY C E+G +GR HYH IL L +G P+ ++ +I E
Sbjct 75 WANQRVHATSLRYFWCREFGKMYGRKHYHVILL-LNKDTWCSLGDFSE--PSSLATMIQE 131
Query 157 CWPFGFH 163
W H
Sbjct 132 AWCSALH 138
> sagm:BSA_2880 Rep protein
Length=248
Score = 40.8 bits (94), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 41/158 (26%), Positives = 71/158 (45%), Gaps = 31/158 (20%)
Query 49 RLDRSLDSAVRAHHESL-LYDRNY------FLTLTYSPEHLPPFGSLIPRDLTLFWKRLR 101
+L R + S A HE + L D N+ F+T+T + E++ + ++ F KRL
Sbjct 45 KLKRLVKSRKNAKHELMRLIDTNFDERLTKFITIT-TKENIQD-RKIFNQEFDKFMKRLN 102
Query 102 -----KRGVSLRYMACGEYGSAFGRPHYHAILFNLPAIELKQIGTTHTRFPTYISNVISE 156
+ ++Y+A E G H H ++FNLP I+ +++ E
Sbjct 103 YNVFHSKKKMIKYVAVLE-KQKRGAYHAHILVFNLPFIKFEKL---------------RE 146
Query 157 CWPFGFHTINSVSFETCAYVARYVTKKI-LGDGKQVYE 193
W G IN V+ ++ RY+TK + G G+++ E
Sbjct 147 LWKLGSIRINKVNVDSIDNRGRYITKYMEKGIGQELIE 184
> btl:BALH_0258 Rep protein
Length=276
Score = 40.0 bits (92), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 28/88 (32%), Positives = 36/88 (41%), Gaps = 22/88 (25%)
Query 103 RGVSLRYMACGEYGSAFGRPHYHAILFNLPAI---ELKQIGTTHTRFPTYISNVISECWP 159
+ +L+Y+A E G HYH I FNLP I ELK I W
Sbjct 131 KKATLKYLATWE-KQKRGSIHYHVIFFNLPFIKNLELKNI------------------WK 171
Query 160 FGFHTINSVSFETCAYVARYVTKKILGD 187
GF IN + ++ RYV+K D
Sbjct 172 HGFVKINKIDVDSKDNRGRYVSKYFSKD 199
> ecz:ECS88_3297 hypothetical protein
Length=246
Score = 39.7 bits (91), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 23/63 (37%), Positives = 29/63 (46%), Gaps = 3/63 (5%)
Query 101 RKRGVSLRYMACGEYGSAFGRPHYHAILFNLPAIELKQIGTTHTRFPTYISNVISECWPF 160
R SLRY C E+G +GR HYH IL L IG P+ ++ +I E W
Sbjct 79 RVHATSLRYFWCREFGKMYGRKHYHVILL-LNKDTWCSIGDFSE--PSSLATMIQEAWCS 135
Query 161 GFH 163
H
Sbjct 136 ALH 138
> ecv:APECO1_3496 hypothetical protein
Length=246
Score = 39.7 bits (91), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 23/63 (37%), Positives = 29/63 (46%), Gaps = 3/63 (5%)
Query 101 RKRGVSLRYMACGEYGSAFGRPHYHAILFNLPAIELKQIGTTHTRFPTYISNVISECWPF 160
R SLRY C E+G +GR HYH IL L IG P+ ++ +I E W
Sbjct 79 RVHATSLRYFWCREFGKMYGRKHYHVILL-LNKDTWCSIGDFSE--PSSLATMIQEAWCS 135
Query 161 GFH 163
H
Sbjct 136 ALH 138
> ecc:c5172 hypothetical protein
Length=246
Score = 39.7 bits (91), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 23/63 (37%), Positives = 29/63 (46%), Gaps = 3/63 (5%)
Query 101 RKRGVSLRYMACGEYGSAFGRPHYHAILFNLPAIELKQIGTTHTRFPTYISNVISECWPF 160
R SLRY C E+G +GR HYH IL L IG P+ ++ +I E W
Sbjct 79 RVHATSLRYFWCREFGKMYGRKHYHVILL-LNKDTWCSIGDFSE--PSSLATMIQEAWCS 135
Query 161 GFH 163
H
Sbjct 136 ALH 138
> cja:CJA_2629 hypothetical protein
Length=172
Score = 38.1 bits (87), Expect = 0.89, Method: Compositional matrix adjust.
Identities = 32/101 (32%), Positives = 42/101 (42%), Gaps = 6/101 (6%)
Query 43 GQCIGCRLDRSLDSAVRAHHESLLYDRNYFLTLTYSPEHLPPFGSLIPRDLTLFW----K 98
G+ I LD+ + L +D + P H+P SLIP L LF K
Sbjct 46 GRMINQTLDQDKRAKALGLSAELRFDLESGEVILRFP-HIPDDASLIPEQLLLFMSHPVK 104
Query 99 RLRKRGVSLRYMACGEY-GSAFGRPHYHAILFNLPAIELKQ 138
R + + LR M G+Y G RP Y L P +EL Q
Sbjct 105 ASRDQPIQLRAMGAGQYHGELAIRPEYAWYLTLYPVVELAQ 145
> fph:Fphi_0992 hypothetical protein
Length=149
Score = 37.7 bits (86), Expect = 0.89, Method: Compositional matrix adjust.
Identities = 30/127 (24%), Positives = 55/127 (43%), Gaps = 12/127 (9%)
Query 127 ILFNLPAIELKQIGTTHTRFPTYISNVISECWPFGFHTINSVSFETCAYVARYVTKKILG 186
IL + ++ + T + IS V++ + G+ +IN + F+ Y A+ +
Sbjct 11 ILLTVISMSVMYADDTPSSTAVPISKVVANIYSQGYPSINKIEFDDGVYKAKVINDA--- 67
Query 187 DGKQVYEKFDPITGEV---DCRVKEFSRWSTKPGIGHDYFQKYW-----RDFYKIDCCLI 238
GK+ Y DP TG+V KE + I +D + D Y+++C
Sbjct 68 -GKEEYLYIDPNTGKVPLPKNGSKEINMSKAIAAIPNDRCKTITSVEDKSDVYEVECVDS 126
Query 239 NNKKFKI 245
+NK+ K+
Sbjct 127 SNKEVKV 133
> rto:RTO_18270 hypothetical protein
Length=253
Score = 38.9 bits (89), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 41/143 (29%), Positives = 58/143 (41%), Gaps = 32/143 (22%)
Query 53 SLDSAVRAHHESLLY--DRNYFLTLTYSPEHLPPFGSLIPRDLTLFWKRLR----KRGVS 106
+++ A A H L Y +YFLTLTY E PP + +D T +LR K +
Sbjct 49 AMNKAETARHRLLEYFGKGDYFLTLTYRVEARPPDMAKAKKDFTNLISKLRTRYKKEQIE 108
Query 107 LRYMACGEYGSAFGRPHYHAILFNLPAIELKQIGTTHTRFPTYISNVISECWPFGFHTIN 166
LR++ E G+ G H H ++ G T + ECWP G I
Sbjct 109 LRWIRNIEKGTK-GAWHVHMVI----------TGCRDT------IRWVEECWPHG--GIY 149
Query 167 SVSFETCAY-------VARYVTK 182
+ E Y +A Y+TK
Sbjct 150 AEQLEKSKYYEEDFSQLASYITK 172
> ena:ECNA114_4547 hypothetical protein
Length=246
Score = 38.5 bits (88), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 22/63 (35%), Positives = 29/63 (46%), Gaps = 3/63 (5%)
Query 101 RKRGVSLRYMACGEYGSAFGRPHYHAILFNLPAIELKQIGTTHTRFPTYISNVISECWPF 160
R SLRY C E+G +GR HYH IL L +G P+ ++ +I E W
Sbjct 79 RVHATSLRYFWCREFGKMYGRKHYHVILL-LNKDTWCSLGDFSE--PSSLATMIQEAWCS 135
Query 161 GFH 163
H
Sbjct 136 ALH 138
> ppa:PAS_chr2-1_0697 Catalytic subunit of DNA polymerase zeta,
which is involved in DNA repair and translesion synthesis
Length=1495
Score = 39.3 bits (90), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 40/164 (24%), Positives = 69/164 (42%), Gaps = 16/164 (10%)
Query 160 FGFHTINSVSFETCAYVARYVTK--KILGDGKQVYEKFDPITGEVDCRVKEFSRWSTKPG 217
+GFH ++S + C +YV K K+L +GK K P + ++ + ++ P
Sbjct 129 YGFHVMHSAFVKICLLNPKYVPKLSKLLSEGKIFGSKIQPFESHIPYTMQFLTDYNLFPC 188
Query 218 IGHDYFQKYWRDFYKIDCCLINNKKFKIPRYYDRLLLRENPDVFEIVKQKRILSAQSY-R 276
Q YWR + N + + RL NP + + +LSA S+ R
Sbjct 189 DWLKIKQWYWR-----SPLIDGNSQLNQDKLNKRLKDYLNPRIINRKNKLNVLSASSFQR 243
Query 277 LT-----PDAQKSRLLVREEVKRLRVERLLR---PFEAQISEYL 312
+T D +L R++++ + RLL E+Q +YL
Sbjct 244 ITNSFFEADINADWILNRDQLRERDLHRLLNLEPDVESQTEKYL 287
> dgi:Desgi_0489 glycyl-radical enzyme activator family protein
Length=311
Score = 38.1 bits (87), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 19/48 (40%), Positives = 27/48 (56%), Gaps = 4/48 (8%)
Query 140 GTTHTRFPTYISNVISECWPFGFHTINSVSFETCAYVARYVTKKILGD 187
G T+ Y+ +++ C +GFH ++ ETC YVA V K ILGD
Sbjct 145 GGEPTQQIVYLRSLLKLCGAYGFH----IALETCGYVAWEVLKSILGD 188
> ecw:EcE24377A_3395 hypothetical protein
Length=243
Score = 37.7 bits (86), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 22/63 (35%), Positives = 29/63 (46%), Gaps = 3/63 (5%)
Query 101 RKRGVSLRYMACGEYGSAFGRPHYHAILFNLPAIELKQIGTTHTRFPTYISNVISECWPF 160
R SLRY C E+G +GR HYH IL L +G P+ ++ +I E W
Sbjct 79 RVHATSLRYFWCREFGKMYGRKHYHVILL-LNKDTWCSLGDFSE--PSSLATMILEAWCS 135
Query 161 GFH 163
H
Sbjct 136 ALH 138
> elo:EC042_3214 hypothetical protein
Length=243
Score = 37.7 bits (86), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 22/63 (35%), Positives = 29/63 (46%), Gaps = 3/63 (5%)
Query 101 RKRGVSLRYMACGEYGSAFGRPHYHAILFNLPAIELKQIGTTHTRFPTYISNVISECWPF 160
R SLRY C E+G +GR HYH IL L +G P+ ++ +I E W
Sbjct 79 RVHATSLRYFWCREFGKMYGRKHYHVILL-LNKDTWCSLGDFSE--PSSLATMILEAWCS 135
Query 161 GFH 163
H
Sbjct 136 ALH 138
> cle:Clole_0725 hypothetical protein
Length=241
Score = 37.4 bits (85), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 35/151 (23%), Positives = 61/151 (40%), Gaps = 29/151 (19%)
Query 51 DRSLDSAVRAHHESLLYDRNYFLTLTYSPEHLPPFGSLIPRDLTLFWKRLRK----RGVS 106
+R + +R + + +Y L L + PE P + + F + LRK + +
Sbjct 47 NRMAEKKLRRKINANFGEGDYHLVLNFRPEERPKTKQEAKKQIENFIRSLRKEYREQNME 106
Query 107 LRYMACGEYGSAFGRPHYHAILFNLPAIELKQIGTTHTRFPTYISNVISECWPFGFHTIN 166
L+Y+ E G G H+H ++ + IS+ W FG IN
Sbjct 107 LKYIHVIEQGKN-GAMHHHLVINEIDP------------------KTISKAWKFGRININ 147
Query 167 SVSFETCAY--VARYV---TKKILGDGKQVY 192
+ ET Y +A Y+ T K++G + +Y
Sbjct 148 PLD-ETGQYAKLASYLIKYTSKVIGTDRAIY 177
> dha:DEHA2D13838g DEHA2D13838p
Length=484
Score = 37.7 bits (86), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 27/84 (32%), Positives = 43/84 (51%), Gaps = 6/84 (7%)
Query 160 FGFHTINSVSFETC-AYVARYVTKKILGDGKQVYEKFDPITGEVDCRVKEFSRWSTKPGI 218
+GF+ +N+ + + + TK+I GD Q + D T D RV +F + S KP +
Sbjct 397 YGFNPMNAPNLSLLQTELEIWETKRIKGDSSQ---QGDSETESDDRRVLDFEKTSMKPYV 453
Query 219 GHDYFQKYWRDFYKIDCCLINNKK 242
DYF K+ D K + + NNK+
Sbjct 454 --DYFAKFVNDLLKDELKVANNKR 475
> yen:YE1686 gpA; phage replication protein
Length=762
Score = 37.7 bits (86), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 49/99 (49%), Gaps = 7/99 (7%)
Query 122 PHYHAILFNLPA-IEL-KQIGTTHTRFPTYISNVISECWPFGFHT--INSVSFETCAYVA 177
PH+H +LF LPA IEL + I T+ R+ E FH I+ Y+A
Sbjct 396 PHWHMLLFMLPADIELARDIFCTYARWEDSEELQSQEALKARFHVVPIDKELGSATGYIA 455
Query 178 RYVTKKILGDGKQVYEKFDPITGE-VDCRVKEFSRWSTK 215
+Y++K I DG + ++ D +G+ + K S W+++
Sbjct 456 KYISKNI--DGYALDDELDDESGKPLKETAKRVSAWASR 492
> dha:DEHA2F16346g DEHA2F16346p
Length=840
Score = 38.1 bits (87), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 23/70 (33%), Positives = 37/70 (53%), Gaps = 9/70 (13%)
Query 155 SECWPFGFH--TINSVSFETCAYVARYVTKKILGDGKQVYEKFDPITGEVDCRVKEFSRW 212
S+ P+G H T+NS +E+C + T+ + G K DP+TG+V+ RV + R
Sbjct 533 SQYTPYGIHKNTLNSKKYESCDWSKISWTRNLRG-------KADPVTGQVNFRVTKGFRD 585
Query 213 STKPGIGHDY 222
+ G+ DY
Sbjct 586 ILRKGVKPDY 595
> lgv:LCGL_0019 hypothetical protein
Length=280
Score = 37.4 bits (85), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 45/195 (23%), Positives = 78/195 (40%), Gaps = 26/195 (13%)
Query 1 MTCYHPITAYWSRTLKTKLGTPAITFKYADADPELGEFQIPCGQCIGCRLDRSLDSAVRA 60
+T +H S+ K K G A + +A E E Q+ S+ +R
Sbjct 20 VTTFHSPQLRSSQEGKHKGGRSADSVISDEAQQERTEKQM-----------FSIKRKIR- 67
Query 61 HHESLLYDRNYFLTLTYSPEHLPPFGSLIPRDLTLFWKRL-RKRGVSLRYMACGEYGSAF 119
H L D YF+TLT P+ I + L W R RK Y+ E+ +
Sbjct 68 -HYILANDFKYFVTLTIDPKKRDSLDYEIAKQTLLKWCRAQRKTKGKFDYIFVPEFHKS- 125
Query 120 GRPHYHAIL-------FNLPAIELKQIGTTHTRFPTYISNVISECWPFGFHTINSVSFE- 171
GR H+H ++ F L + G R + + + W +GF + + +
Sbjct 126 GRVHFHGVIGQPEYRSFTLTEATNPKTGEILIRHNRQVLEL--KEWRYGFTDVTPIEDKE 183
Query 172 -TCAYVARYVTKKIL 185
T +Y+ +Y++K+++
Sbjct 184 RTSSYMTKYISKELM 198
> lgr:LCGT_0019 hypothetical protein
Length=280
Score = 37.4 bits (85), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 45/195 (23%), Positives = 78/195 (40%), Gaps = 26/195 (13%)
Query 1 MTCYHPITAYWSRTLKTKLGTPAITFKYADADPELGEFQIPCGQCIGCRLDRSLDSAVRA 60
+T +H S+ K K G A + +A E E Q+ S+ +R
Sbjct 20 VTTFHSPQLRSSQEGKHKGGRSADSVISDEAQQERTEKQM-----------FSIKRKIR- 67
Query 61 HHESLLYDRNYFLTLTYSPEHLPPFGSLIPRDLTLFWKRL-RKRGVSLRYMACGEYGSAF 119
H L D YF+TLT P+ I + L W R RK Y+ E+ +
Sbjct 68 -HYILANDFKYFVTLTIDPKKRDSLDYEIAKQTLLKWCRAQRKTKGKFDYIFVPEFHKS- 125
Query 120 GRPHYHAIL-------FNLPAIELKQIGTTHTRFPTYISNVISECWPFGFHTINSVSFE- 171
GR H+H ++ F L + G R + + + W +GF + + +
Sbjct 126 GRVHFHGVIGQPEYRSFTLTEATNPKTGEILIRHNRQVLEL--KEWRYGFTDVTPIEDKE 183
Query 172 -TCAYVARYVTKKIL 185
T +Y+ +Y++K+++
Sbjct 184 RTSSYMTKYISKELM 198
> ypb:YPTS_3589 replication gene A
Length=695
Score = 37.4 bits (85), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 28/101 (28%), Positives = 46/101 (46%), Gaps = 7/101 (7%)
Query 120 GRPHYHAILFNLP--AIELKQIGTTHTRFPTYISNVISECWPFGFHT--INSVSFETCAY 175
G PH+H +LF LP +++ I + R + E FH I+S Y
Sbjct 394 GTPHWHVLLFMLPQHVDQVRDILCYYARLEDSETLQSPEALKARFHAEPIDSAKGSATGY 453
Query 176 VARYVTKKILGDGKQVYEKFDPIT-GEVDCRVKEFSRWSTK 215
+A+Y++K I DG + E+ D T G K + W+++
Sbjct 454 IAKYISKNI--DGYALDEEEDGETGGNARDMAKAVTAWASR 492
> mag:amb2284 response regulator containing CheY-like receiver
Length=479
Score = 36.6 bits (83), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 33/73 (45%), Gaps = 16/73 (22%)
Query 51 DRSLDSAVRAHHESLLYDRNYFLTLTYSPEHLPPFGSLIPRDLTLFWKRLRKRGVSLRYM 110
+R+L+ VR H + R+ F L P HLPP P D+ L +RL
Sbjct 305 NRNLEEEVREKH----FRRDLFYRLAAFPIHLPPLRER-PMDVPLLAERL---------- 349
Query 111 ACGEYGSAFGRPH 123
GE G FGRP+
Sbjct 350 -LGETGKGFGRPN 361
> aoe:Clos_1826 hypothetical protein
Length=344
Score = 36.6 bits (83), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 32/121 (26%), Positives = 54/121 (45%), Gaps = 28/121 (23%)
Query 72 FLTLTYSPEHLPPFGSLIPRDLTLFWKRL-----RKRGVSLRYMACGEYGSAFGRPHYHA 126
FLTLT++ E++ + + T F KRL + L+Y+ E+ G H+H
Sbjct 98 FLTLTFA-ENITDLQT-ANAEFTTFNKRLSYHLYKTNKNVLKYICIPEFQKR-GAVHFHV 154
Query 127 ILFNLPAIELKQIGTTHTRFPTYISNVISECWPFGFHTINSVSFET-----CAYVARYVT 181
+ FNLP + +K+ ISE W G+ I ++ + YV +Y++
Sbjct 155 LYFNLPYVNIKK---------------ISEVWGHGYAFIEGITEKQNIEDFAKYVCKYMS 199
Query 182 K 182
K
Sbjct 200 K 200
> bom:102267479 FSTL4; follistatin-like 4
Length=843
Score = 36.6 bits (83), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 37/133 (28%), Positives = 58/133 (44%), Gaps = 7/133 (5%)
Query 182 KKILGDGKQVYEKFDPITGEVDCRVKEFSRWSTKPGIGHD--YFQKYWRDFYKIDCCLIN 239
KK+ G G + + TGE +CR E R S P G D ++ + + +++ C L
Sbjct 67 KKLCGRGSRCM--LNRKTGEPECRCLEACRPSYVPVCGSDGRVYENHC-ELHRVACLL-- 121
Query 240 NKKFKIPRYYDRLLLRENPDVFEIVKQKRILSAQSYRLTPDAQKSRLLVREEVKRLRVER 299
KK + D L + V + + K++L A RL P + KRL VE
Sbjct 122 GKKIVMVHNKDCFLKGDPCTVADYARLKKVLLALQSRLQPLHEGDSRQDPASQKRLLVES 181
Query 300 LLRPFEAQISEYL 312
L + F+A +L
Sbjct 182 LFKDFDADGDGHL 194
> xcv:XCV2471 filamentous phage phiLf replication initiation protein
II
Length=346
Score = 36.2 bits (82), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 43/156 (28%), Positives = 58/156 (37%), Gaps = 38/156 (24%)
Query 44 QCIGCRLDRSLDSAVRAHHESL----LYDRNYFLTLTYSPEHLPPFGSLIPRDLTLFWKR 99
Q RL +S+ + R H + YFLTLTY PRD++ +KR
Sbjct 127 QARAQRLRKSVITGARLHDQEAKKGSFRGAWYFLTLTYRDGSDSS-----PRDVSELFKR 181
Query 100 LR------KRG------VSLRYMACGEYGSAFGRPHYHAILFNLPAIELKQIGTTHTRFP 147
+R K G S RY+ GE F RPHYH +L+ +
Sbjct 182 MRGHFNRLKSGRARWNRESFRYVWVGELTQRF-RPHYHVMLWVPTGM------------- 227
Query 148 TYISNVISEC-WPFGFHTINSVSFETCAYVARYVTK 182
Y V WP G I Y+A+Y +K
Sbjct 228 -YFGKVDQRGWWPHGTTQIEKAR-NCVGYLAKYASK 261
> fnl:M973_09525 peptidase
Length=149
Score = 35.0 bits (79), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 30/113 (27%), Positives = 51/113 (45%), Gaps = 16/113 (14%)
Query 142 THTRFPTYISNVISECWPFGFHTINSVSFETCAYVARYVTKKILGD-GKQVYEKFDPITG 200
+ T P IS V++ + G+ +IN + F+ Y A K++ D GK+ Y DP TG
Sbjct 28 SSTAVP--ISKVVANIYSQGYPSINKIEFDDGVYKA-----KVINDVGKEEYLYIDPNTG 80
Query 201 EVDCRVKEFSRWSTKPGIG---HDYFQKYW-----RDFYKIDCCLINNKKFKI 245
+V K ++ + I D + D Y+++C NK+ K+
Sbjct 81 KVPLPKKGSTQINMSDAIAAVPSDRCKTIISVEDKSDVYEVECVDTANKEVKV 133
> fna:OOM_0578 hypothetical protein
Length=149
Score = 35.0 bits (79), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 30/113 (27%), Positives = 51/113 (45%), Gaps = 16/113 (14%)
Query 142 THTRFPTYISNVISECWPFGFHTINSVSFETCAYVARYVTKKILGD-GKQVYEKFDPITG 200
+ T P IS V++ + G+ +IN + F+ Y A K++ D GK+ Y DP TG
Sbjct 28 SSTAVP--ISKVVANIYSQGYPSINKIEFDDGVYKA-----KVINDVGKEEYLYIDPNTG 80
Query 201 EVDCRVKEFSRWSTKPGIG---HDYFQKYW-----RDFYKIDCCLINNKKFKI 245
+V K ++ + I D + D Y+++C NK+ K+
Sbjct 81 KVPLPKKGSTQINMSDAIAAVPSDRCKTIISVEDKSDVYEVECVDTANKEVKV 133
> xcc:XCC2058 gII; replication initiation protein
Length=346
Score = 36.2 bits (82), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 33/102 (32%), Positives = 44/102 (43%), Gaps = 22/102 (22%)
Query 44 QCIGCRLDRSLDSAVRAHHESL----LYDRNYFLTLTYSPEHLPPFGSLIPRDLTLFWKR 99
Q RL +S+ + R H + YFLTLTY PRD++ +KR
Sbjct 127 QARAQRLRKSVITGARLHDQEAKKGSFRGAWYFLTLTYRDGS-----DSSPRDVSELFKR 181
Query 100 LR------KRG------VSLRYMACGEYGSAFGRPHYHAILF 129
+R K G S RY+ GE F RPHYH +L+
Sbjct 182 MRGHFNRLKSGRARWNRESFRYVWVGELTQRF-RPHYHVMLW 222
> mjd:JDM601_4242 tesB2; acyl-CoA thioesterase
Length=323
Score = 36.2 bits (82), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 39/99 (39%), Gaps = 9/99 (9%)
Query 6 PITAYWSRTLKTKLGT---------PAITFKYADADPELGEFQIPCGQCIGCRLDRSLDS 56
P+ +W++ +L P + + ADA LG Q + + RL R +D
Sbjct 159 PLLQHWAQQAPRELAGSASTWVDTPPPLEMRIADAPIFLGGTQAGGARSLWMRLPRPIDG 218
Query 57 AVRAHHESLLYDRNYFLTLTYSPEHLPPFGSLIPRDLTL 95
AH L Y +Y L T H P G R LTL
Sbjct 219 GPGAHAAMLAYASDYLLVDTAFRAHPEPVGYRTHRGLTL 257
> mch:Mchl_3336 PAS/PAC sensor hybrid histidine kinase
Length=1065
Score = 36.6 bits (83), Expect = 9.0, Method: Composition-based stats.
Identities = 21/51 (41%), Positives = 24/51 (47%), Gaps = 12/51 (24%)
Query 187 DGKQVYE-------KFDPITGEVDCRVKEFSRWSTKPGIGHDYFQKYWRDF 230
D KQV E KF I +D V WST+P HDYF + W DF
Sbjct 412 DRKQVEERLRESEAKFQAIANSIDQMV-----WSTRPDGHHDYFNQRWYDF 457
> xcb:XC_2107 replication initiation protein
Length=346
Score = 36.2 bits (82), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 33/102 (32%), Positives = 44/102 (43%), Gaps = 22/102 (22%)
Query 44 QCIGCRLDRSLDSAVRAHHESL----LYDRNYFLTLTYSPEHLPPFGSLIPRDLTLFWKR 99
Q RL +S+ + R H + YFLTLTY PRD++ +KR
Sbjct 127 QARAQRLRKSVITGARLHDQEAKKGSFRGAWYFLTLTYRDGSDSS-----PRDVSELFKR 181
Query 100 LR------KRG------VSLRYMACGEYGSAFGRPHYHAILF 129
+R K G S RY+ GE F RPHYH +L+
Sbjct 182 MRGHFNRLKSGRARWNRESFRYVWVGELTQRF-RPHYHVMLW 222
> xcc:XCC2075 gII; replication initiation protein
Length=346
Score = 36.2 bits (82), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 33/102 (32%), Positives = 44/102 (43%), Gaps = 22/102 (22%)
Query 44 QCIGCRLDRSLDSAVRAHHESL----LYDRNYFLTLTYSPEHLPPFGSLIPRDLTLFWKR 99
Q RL +S+ + R H + YFLTLTY PRD++ +KR
Sbjct 127 QARAQRLRKSVITGARLHDQEAKKGSFRGAWYFLTLTYRDGSDSS-----PRDVSELFKR 181
Query 100 LR------KRG------VSLRYMACGEYGSAFGRPHYHAILF 129
+R K G S RY+ GE F RPHYH +L+
Sbjct 182 MRGHFNRLKSGRARWNRESFRYVWVGELTQRF-RPHYHVMLW 222
> nvi:100679173 G-protein coupled receptor Mth2-like
Length=430
Score = 36.2 bits (82), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 23/88 (26%), Positives = 40/88 (45%), Gaps = 7/88 (8%)
Query 225 KYWRDFYKIDCCLINNKKFKIPRYYDRLLLRENPDVFEIVKQKRILSAQSYRLTPDA--- 281
K W ++ ++DCC + +FK Y ++ +FE++ + Q Y + PD
Sbjct 312 KLWHEYRELDCCKLKVLRFKCTMYAKLAVMMGFTWIFEVISFASGETNQFYWILPDVLNV 371
Query 282 -QKSRLLVREEVKRLRVERLL---RPFE 305
Q + + V R RV +LL +PF
Sbjct 372 LQGIFIFILLVVSRKRVRKLLAKRKPFN 399
> xcp:XCR_2349 filamentous phage phiLf replication initiation
protein II
Length=346
Score = 35.8 bits (81), Expect = 10.0, Method: Compositional matrix adjust.
Identities = 33/102 (32%), Positives = 44/102 (43%), Gaps = 22/102 (22%)
Query 44 QCIGCRLDRSLDSAVRAHHESL----LYDRNYFLTLTYSPEHLPPFGSLIPRDLTLFWKR 99
Q RL +S+ + R H + YFLTLTY PRD++ +KR
Sbjct 127 QARAQRLRKSVITGARLHDQEAKKGSFRGAWYFLTLTYRDGS-----DSSPRDVSELFKR 181
Query 100 LR------KRG------VSLRYMACGEYGSAFGRPHYHAILF 129
+R K G S RY+ GE F RPHYH +L+
Sbjct 182 MRGHFNRLKSGRARWNRESFRYVWVGELTQRF-RPHYHVMLW 222
Lambda K H a alpha
0.325 0.141 0.451 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 553088382528