bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
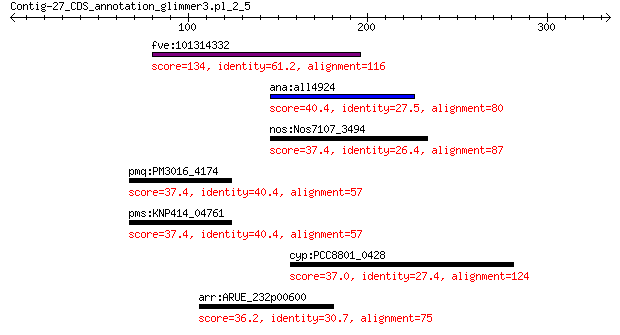
Query= Contig-27_CDS_annotation_glimmer3.pl_2_5
Length=334
Score E
Sequences producing significant alignments: (Bits) Value
fve:101314332 capsid protein VP1-like 134 6e-33
ana:all4924 ferrichrome iron receptor 40.4 0.48
nos:Nos7107_3494 TonB-dependent siderophore receptor 37.4 4.5
pmq:PM3016_4174 hypothetical protein 37.4 4.6
pms:KNP414_04761 yesN17; hypothetical protein 37.4 4.7
cyp:PCC8801_0428 response regulator receiver modulated PAS/PAC... 37.0 5.7
arr:ARUE_232p00600 Mn-or Fe-dependent transcriptional regulator 36.2 7.2
> fve:101314332 capsid protein VP1-like
Length=421
Score = 134 bits (338), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 71/118 (60%), Positives = 83/118 (70%), Gaps = 2/118 (2%)
Query 80 TAATINQLRMAFQIQKLYEKDARGGTRYIEILKSHFGVTSPDARLQRPEYLGGNRIPVNI 139
TAATINQLR +FQIQKL E+DARGGTRY EI++SHFGV SPDARLQRPEYLGG P+NI
Sbjct 304 TAATINQLRQSFQIQKLLERDARGGTRYTEIIRSHFGVASPDARLQRPEYLGGGSTPINI 363
Query 140 NQV--VQSSATQSSGTPLGDTAAFSVTTDVHGDFIKSFVEHGFVIGIMVARYDHTYQQ 195
+ + Q + TP G+ AAF F +SFVEHG VIG++ R D TYQQ
Sbjct 364 APIAQTGGTGAQGTTTPQGNLAAFGTYMAKGHGFSQSFVEHGHVIGLVSVRADLTYQQ 421
> ana:all4924 ferrichrome iron receptor
Length=858
Score = 40.4 bits (93), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 22/80 (28%), Positives = 44/80 (55%), Gaps = 6/80 (8%)
Query 146 SATQSSGTPLGDTAAFSVTTDVHGDFIKSFVEHGFVIGIMVARYDHTYQQGLERFWSRRD 205
+A +S+ + D+ +++TTD+ G F +EH + G+ ++R+D+ G++ +
Sbjct 492 TANRSTYSSESDSNIYNLTTDISGRFFTGSIEHQLLFGVNMSRFDNFNNFGID--LAELT 549
Query 206 RLDYYFPVFANIGEQPILNK 225
LD Y PV+ QPI+ +
Sbjct 550 PLDIYNPVY----RQPIVGR 565
> nos:Nos7107_3494 TonB-dependent siderophore receptor
Length=896
Score = 37.4 bits (85), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 23/89 (26%), Positives = 46/89 (52%), Gaps = 8/89 (9%)
Query 146 SATQSSGTPLGDTAAFSVTTDVHGDFIKSFVEHGFVIGIMVARYDHTYQQGLERFWSRRD 205
+A +S+ D+ +++TTD+ G F +EH + G+ ++R+D+ G++ +
Sbjct 530 TANRSTYNSESDSNIYNLTTDISGRFSTGSIEHQLLFGVNMSRFDNFNNFGID--LALLS 587
Query 206 RLDYYFPVFANIGEQPILNK--EIYAQGT 232
LD Y P + QPI+ + +Y G+
Sbjct 588 PLDIYNPAY----RQPIVGRIDAVYEDGS 612
> pmq:PM3016_4174 hypothetical protein
Length=550
Score = 37.4 bits (85), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 23/68 (34%), Positives = 39/68 (57%), Gaps = 11/68 (16%)
Query 67 YLDNLWAIQSGNVTAATINQLRM---AFQIQ----KLYEKDARGGTRY----IEILKSHF 115
YL ++ +++G + TI +LRM A+ ++ K+YE AR G +Y I++ K H+
Sbjct 476 YLSRIYKLETGESISDTIYKLRMEQAAYYLRSTELKIYEITARLGYQYPPYFIKVFKQHY 535
Query 116 GVTSPDAR 123
GVT + R
Sbjct 536 GVTPQEYR 543
> pms:KNP414_04761 yesN17; hypothetical protein
Length=550
Score = 37.4 bits (85), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 23/68 (34%), Positives = 39/68 (57%), Gaps = 11/68 (16%)
Query 67 YLDNLWAIQSGNVTAATINQLRM---AFQIQ----KLYEKDARGGTRY----IEILKSHF 115
YL ++ +++G + TI +LRM A+ ++ K+YE AR G +Y I++ K H+
Sbjct 476 YLSRIYKLETGESISDTIYKLRMEQAAYYLRSTELKIYEITARLGYQYPPYFIKVFKQHY 535
Query 116 GVTSPDAR 123
GVT + R
Sbjct 536 GVTPQEYR 543
> cyp:PCC8801_0428 response regulator receiver modulated PAS/PAC
sensor-containing diguanylate cyclase
Length=455
Score = 37.0 bits (84), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 34/124 (27%), Positives = 55/124 (44%), Gaps = 19/124 (15%)
Query 157 DTAAFSVTTDVHGDFIKSFVEHGFVIGIMVARYDHTYQQGLERFWSRRDRLDYYFPVFAN 216
+T A + T+ +G IK FVE I + +A Y H QQ L + + F + +
Sbjct 88 ETVARAKLTNPYGYLIKPFVEQDLRITLEIALYKHQIQQTLIEYKQ------WLFTILQS 141
Query 217 IGEQPILNKEIYAQGTVQDNEVFGYQEAWADYRYKPSRVAGEMRSKAPTSLDVWHLADEY 276
+G+ +L +I+ Q T N + + W+ E R K T +V+ L DE
Sbjct 142 VGDG-VLTTDIHGQITYL-NPMAEHLTGWS---------LAEARGKPVT--EVFKLIDEV 188
Query 277 TQLP 280
T+ P
Sbjct 189 TRQP 192
> arr:ARUE_232p00600 Mn-or Fe-dependent transcriptional regulator
Length=232
Score = 36.2 bits (82), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 23/75 (31%), Positives = 32/75 (43%), Gaps = 8/75 (11%)
Query 106 RYIEILKSHFGVTSPDARLQRPEYLGGNRIPVNINQVVQSSATQSSGTPLGDTAAFSVTT 165
+ IE + H G + D G+ IP Q Q +ATQ S P G T A + +
Sbjct 114 KLIERIDQHLGHPTRDPH--------GDPIPSPAGQQHQPAATQLSAAPAGSTVAVARIS 165
Query 166 DVHGDFIKSFVEHGF 180
D D ++ F E G
Sbjct 166 DADPDMLRYFTELGL 180
Lambda K H a alpha
0.318 0.133 0.404 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 606891589622