bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-24_CDS_annotation_glimmer3.pl_2_6
Length=135
Score E
Sequences producing significant alignments: (Bits) Value
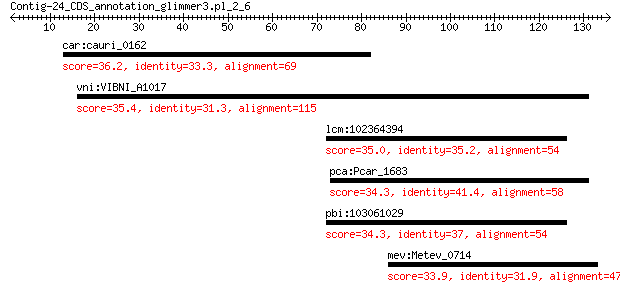
car:cauri_0162 hypothetical protein 36.2 1.7
vni:VIBNI_A1017 putative RNA polymerase sigma-70 factor 35.4 2.1
lcm:102364394 NELFA; negative elongation factor complex member A 35.0 3.3
pca:Pcar_1683 pulM-1; type II secretion system ATPase PulM 34.3 6.2
pbi:103061029 NELFA; negative elongation factor complex member A 34.3 7.3
mev:Metev_0714 hypothetical protein 33.9 7.7
> car:cauri_0162 hypothetical protein
Length=1221
Score = 36.2 bits (82), Expect = 1.7, Method: Composition-based stats.
Identities = 23/71 (32%), Positives = 36/71 (51%), Gaps = 3/71 (4%)
Query 13 SSVSGSDSIISVLISNYPLANGGYLVSFGHDEPDGSFKNYDPVYSLDFESSKL--SKFLE 70
+S SD+++S ++N P +GG FG D PDG+ P SL + + +F E
Sbjct 310 ASTKASDNLVSYALTNTPAGDGGRRF-FGWDRPDGTPWGRLPWGSLGYAQVSMGRDRFAE 368
Query 71 FSSIFLPRGCY 81
+SS L R +
Sbjct 369 YSSQRLARSAF 379
> vni:VIBNI_A1017 putative RNA polymerase sigma-70 factor
Length=220
Score = 35.4 bits (80), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 36/127 (28%), Positives = 58/127 (46%), Gaps = 17/127 (13%)
Query 16 SGSDSIISVLISNYPL---ANGGYLVSFGHDEPDGS---FKNYDPVYS------LDFESS 63
+G S ++V+ N L L FG DEP S F +PV + L+ E
Sbjct 73 TGYSSWLTVITRNVALNRLRTDQRLEFFGDDEPVSSCPTFSVSEPVMNQNILQLLEKEID 132
Query 64 KLSKFLEFSSIFLPRGCYYLPEMELAGFIRSLSSDVSFFEMKLLPASAQLQGLLLVKVDE 123
+L + + ++F+ R + +E A SL DV+ + + A QLQ L+ ++
Sbjct 133 QLP--VMYRTVFVMRSVQGMTSLETAD---SLGLDVNVIKTRYRRARLQLQSQLIAHMER 187
Query 124 ESLSKYE 130
ES+S YE
Sbjct 188 ESMSLYE 194
> lcm:102364394 NELFA; negative elongation factor complex member
A
Length=394
Score = 35.0 bits (79), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 27/54 (50%), Gaps = 0/54 (0%)
Query 72 SSIFLPRGCYYLPEMELAGFIRSLSSDVSFFEMKLLPASAQLQGLLLVKVDEES 125
+S LP C YL + L + L+ V F++K P SA L+ LL K E +
Sbjct 9 ASAMLPLECQYLNKNALTTLVGPLTPPVKHFQLKRKPKSATLRAELLQKSTETA 62
> pca:Pcar_1683 pulM-1; type II secretion system ATPase PulM
Length=295
Score = 34.3 bits (77), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 24/75 (32%), Positives = 40/75 (53%), Gaps = 17/75 (23%)
Query 73 SIFLPRGCYYLPEMELAGFIRSL--SSDVSFFEM-KLLPASAQLQ--------------G 115
++ LP C Y+ M++AGF RS S +V +++ KLLP A L
Sbjct 78 ALSLPDRCGYVMTMDVAGFTRSRKESREVVAWQLRKLLPGIADLHFDYQVLKRTGGGKAH 137
Query 116 LLLVKVDEESLSKYE 130
LL+V +++++L +YE
Sbjct 138 LLVVAMEKQALDRYE 152
> pbi:103061029 NELFA; negative elongation factor complex member
A
Length=529
Score = 34.3 bits (77), Expect = 7.3, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 27/54 (50%), Gaps = 0/54 (0%)
Query 72 SSIFLPRGCYYLPEMELAGFIRSLSSDVSFFEMKLLPASAQLQGLLLVKVDEES 125
SS+ LP C YL + L L+ V F++K P SA L+ LL K E +
Sbjct 144 SSVMLPLECQYLNKNALTTLAGPLTPPVKHFQLKRKPKSATLRAELLQKSTETA 197
> mev:Metev_0714 hypothetical protein
Length=490
Score = 33.9 bits (76), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 15/47 (32%), Positives = 32/47 (68%), Gaps = 0/47 (0%)
Query 86 MELAGFIRSLSSDVSFFEMKLLPASAQLQGLLLVKVDEESLSKYEQE 132
ME+ I+++++D++ F+ ++L + +Q +L KVD++ L+K E E
Sbjct 352 MEMDEKIQNMNNDLTSFKQEILESLENIQSVLENKVDDDKLTKIEDE 398
Lambda K H a alpha
0.313 0.133 0.364 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 125245300132