bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-18_CDS_annotation_glimmer3.pl_2_7
Length=346
Score E
Sequences producing significant alignments: (Bits) Value
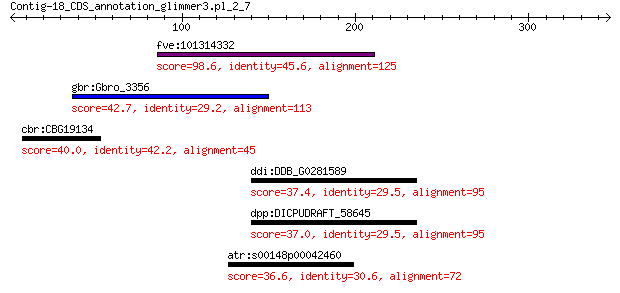
fve:101314332 capsid protein VP1-like 98.6 2e-20
gbr:Gbro_3356 hypothetical protein 42.7 0.035
cbr:CBG19134 Hypothetical protein CBG19134 40.0 0.41
ddi:DDB_G0281589 COBW domain-containing protein 37.4 4.1
dpp:DICPUDRAFT_58645 hypothetical protein 37.0 5.1
atr:s00148p00042460 AMTR_s00148p00042460; hypothetical protein 36.6 9.6
> fve:101314332 capsid protein VP1-like
Length=421
Score = 98.6 bits (244), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 57/126 (45%), Positives = 71/126 (56%), Gaps = 1/126 (1%)
Query 86 LGTDLSNIEAATINQLRQAFAVQHYYEALARGGSRYREQVRALFGVSISDKTVQIPEYLG 145
L DLS AATINQLRQ+F +Q E ARGG+RY E +R+ FGV+ D +Q PEYLG
Sbjct 296 LYADLSAATAATINQLRQSFQIQKLLERDARGGTRYTEIIRSHFGVASPDARLQRPEYLG 355
Query 146 GGRYHVNMNQI-VQTSGQESNYGTPIGETGAMSVTPINESSFTKSFEEHGFVIGVMCVRH 204
GG +N+ I TP G A F++SF EHG VIG++ VR
Sbjct 356 GGSTPINIAPIAQTGGTGAQGTTTPQGNLAAFGTYMAKGHGFSQSFVEHGHVIGLVSVRA 415
Query 205 DHSYQQ 210
D +YQQ
Sbjct 416 DLTYQQ 421
> gbr:Gbro_3356 hypothetical protein
Length=194
Score = 42.7 bits (99), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 33/127 (26%), Positives = 56/127 (44%), Gaps = 18/127 (14%)
Query 37 IVNHTNISFTKEGTKFSVNKNNNGNTAPLVNGQYI---QTMSQDDANFFDAWLGTD---- 89
+V H I+ T + T F+ + + TAP G + +++ A DAW D
Sbjct 41 VVGHI-IAVTDKFTSFARAETDTPRTAPAPRGDPAPLRRALAESVAASADAWPSCDGSRR 99
Query 90 -------LSNIEAATINQLRQAFAVQHYYEALARGGSRYREQVRALFGVSISDKTVQIPE 142
S ++AA+IN + A+ H ++ A G YR RA+ + + + PE
Sbjct 100 CRLPFGTFSAVQAASINMMD---ALVHAWDIAAANGIDYRMPSRAIGPAIVIARRLVTPE 156
Query 143 YLGGGRY 149
+ GG+Y
Sbjct 157 AVAGGQY 163
> cbr:CBG19134 Hypothetical protein CBG19134
Length=257
Score = 40.0 bits (92), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 19/45 (42%), Positives = 26/45 (58%), Gaps = 0/45 (0%)
Query 8 NAPLRAYSEKDLNNRKIGTGFFNNEYNTGIVNHTNISFTKEGTKF 52
++PL S + +N KI F+N EY T H +I F+KEG KF
Sbjct 213 DSPLVLVSSRVVNQDKIDLVFYNPEYKTCNFKHFSIPFSKEGVKF 257
> ddi:DDB_G0281589 COBW domain-containing protein
Length=396
Score = 37.4 bits (85), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 44/100 (44%), Gaps = 9/100 (9%)
Query 140 IPEYLGGGR-----YHVNMNQIVQTSGQESNYGTPIGETGAMSVTPINESSFTKSFEEHG 194
I +LG G+ Y +N N + + ++ YG IG AM V +S+ K E
Sbjct 18 ITGFLGSGKTTFLNYILNENHGKKIAVIQNEYGQSIGIETAMIV----DSNGEKIQEWLE 73
Query 195 FVIGVMCVRHDHSYQQGLERFWSRSDRLDYYFPQFANLGE 234
F G +C + Q +E RSD+ DY + +G+
Sbjct 74 FPNGCICCTVKDDFLQSIEDLLKRSDKFDYILIESTGMGD 113
> dpp:DICPUDRAFT_58645 hypothetical protein
Length=388
Score = 37.0 bits (84), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 44/100 (44%), Gaps = 9/100 (9%)
Query 140 IPEYLGGGR-----YHVNMNQIVQTSGQESNYGTPIGETGAMSVTPINESSFTKSFEEHG 194
I +LG G+ Y +N N + + ++ YG IG AM V +S+ K E
Sbjct 26 ITGFLGSGKTTFLNYILNENHGKKIAVIQNEYGQSIGIETAMIV----DSNGEKVQEWLE 81
Query 195 FVIGVMCVRHDHSYQQGLERFWSRSDRLDYYFPQFANLGE 234
F G +C + Q +E RSD+ DY + +G+
Sbjct 82 FPNGCICCTVKDDFLQSIEDLLKRSDKFDYILIESTGMGD 121
> atr:s00148p00042460 AMTR_s00148p00042460; hypothetical protein
Length=929
Score = 36.6 bits (83), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 35/73 (48%), Gaps = 1/73 (1%)
Query 127 ALFGVSISDKTVQ-IPEYLGGGRYHVNMNQIVQTSGQESNYGTPIGETGAMSVTPINESS 185
ALFG++++ + IP + VN+ +V+T+G++ P+ SVTP
Sbjct 664 ALFGITLAFVVAELIPPFTEDVSRAVNVLHVVETTGKQGGKQNPLSYVSLSSVTPGKLKK 723
Query 186 FTKSFEEHGFVIG 198
S E GFV G
Sbjct 724 EVASLENEGFVCG 736
Lambda K H a alpha
0.316 0.132 0.392 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 644431275578