bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-17_CDS_annotation_glimmer3.pl_2_4
Length=313
Score E
Sequences producing significant alignments: (Bits) Value
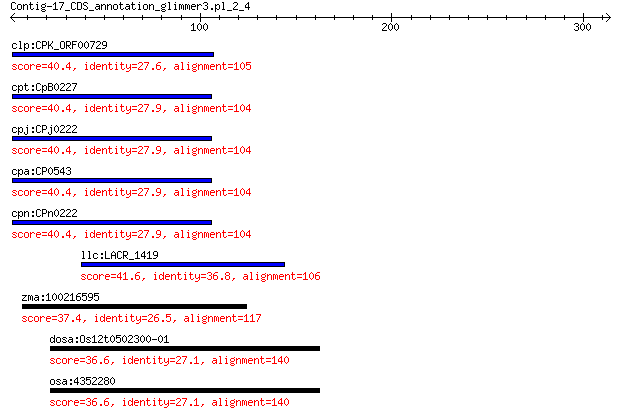
clp:CPK_ORF00729 hypothetical protein 40.4 0.073
cpt:CpB0227 hypothetical protein 40.4 0.077
cpj:CPj0222 hypothetical protein 40.4 0.077
cpa:CP0543 hypothetical protein 40.4 0.077
cpn:CPn0222 hypothetical protein 40.4 0.077
llc:LACR_1419 hypothetical protein 41.6 0.12
zma:100216595 TIDP3618; LOC100216595 37.4 3.3
dosa:Os12t0502300-01 Os12g0502300; Similar to Cyclin A-like pr... 36.6 7.7
osa:4352280 Os12g0502300 36.6 7.7
> clp:CPK_ORF00729 hypothetical protein
Length=121
Score = 40.4 bits (93), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 48/106 (45%), Gaps = 11/106 (10%)
Query 2 RYVTAACGDCYECRKQKQRQWMVRMSEENRQTPNAYFLTLTIDDKSYKQIKQKYNLKDNN 61
R+V C C CR Q + W R E FLTLT DDK +L
Sbjct 18 RWVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDK---------HLPQYG 68
Query 62 DIATKAIRLCLERVR-KLTGKSVKHWFITELGHEKTERLHLHGIVW 106
+ ++L L+R+R +++ ++++ E G K +R H H +++
Sbjct 69 SLVKLHLQLFLKRLRDRISPHKIRYFGCGEYG-TKLQRPHYHLLIF 113
> cpt:CpB0227 hypothetical protein
Length=113
Score = 40.4 bits (93), Expect = 0.077, Method: Compositional matrix adjust.
Identities = 29/104 (28%), Positives = 42/104 (40%), Gaps = 9/104 (9%)
Query 2 RYVTAACGDCYECRKQKQRQWMVRMSEENRQTPNAYFLTLTIDDKSYKQIKQKYNLKDNN 61
R+V C C CR Q + W R E FLTLT DDK +L
Sbjct 18 RWVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDK---------HLPQYG 68
Query 62 DIATKAIRLCLERVRKLTGKSVKHWFITELGHEKTERLHLHGIV 105
+ ++L L+R+RK+ +F K +R H H ++
Sbjct 69 SLVKLHLQLFLKRLRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpj:CPj0222 hypothetical protein
Length=113
Score = 40.4 bits (93), Expect = 0.077, Method: Compositional matrix adjust.
Identities = 29/104 (28%), Positives = 42/104 (40%), Gaps = 9/104 (9%)
Query 2 RYVTAACGDCYECRKQKQRQWMVRMSEENRQTPNAYFLTLTIDDKSYKQIKQKYNLKDNN 61
R+V C C CR Q + W R E FLTLT DDK +L
Sbjct 18 RWVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDK---------HLPQYG 68
Query 62 DIATKAIRLCLERVRKLTGKSVKHWFITELGHEKTERLHLHGIV 105
+ ++L L+R+RK+ +F K +R H H ++
Sbjct 69 SLVKLHLQLFLKRLRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpa:CP0543 hypothetical protein
Length=113
Score = 40.4 bits (93), Expect = 0.077, Method: Compositional matrix adjust.
Identities = 29/104 (28%), Positives = 42/104 (40%), Gaps = 9/104 (9%)
Query 2 RYVTAACGDCYECRKQKQRQWMVRMSEENRQTPNAYFLTLTIDDKSYKQIKQKYNLKDNN 61
R+V C C CR Q + W R E FLTLT DDK +L
Sbjct 18 RWVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDK---------HLPQYG 68
Query 62 DIATKAIRLCLERVRKLTGKSVKHWFITELGHEKTERLHLHGIV 105
+ ++L L+R+RK+ +F K +R H H ++
Sbjct 69 SLVKLHLQLFLKRLRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpn:CPn0222 hypothetical protein
Length=113
Score = 40.4 bits (93), Expect = 0.077, Method: Compositional matrix adjust.
Identities = 29/104 (28%), Positives = 42/104 (40%), Gaps = 9/104 (9%)
Query 2 RYVTAACGDCYECRKQKQRQWMVRMSEENRQTPNAYFLTLTIDDKSYKQIKQKYNLKDNN 61
R+V C C CR Q + W R E FLTLT DDK +L
Sbjct 18 RWVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDK---------HLPQYG 68
Query 62 DIATKAIRLCLERVRKLTGKSVKHWFITELGHEKTERLHLHGIV 105
+ ++L L+R+RK+ +F K +R H H ++
Sbjct 69 SLVKLHLQLFLKRLRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> llc:LACR_1419 hypothetical protein
Length=284
Score = 41.6 bits (96), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 39/126 (31%), Positives = 57/126 (45%), Gaps = 30/126 (24%)
Query 38 FLTLTIDDKSYKQIKQKYNLKDNNDIATKAIRLCLERVRKLTGKSVKHWFITELGHEKTE 97
F TLT DD+ K N + N A K ++ L+ +R+ G+ F+ EL H+ +E
Sbjct 72 FWTLTFDDR-------KVNAR-NYQYARKRLQAWLKYMRETYGR-FGFLFVPEL-HKSSE 121
Query 98 RLHLHGIVWGLG-----------------NGEKVTN--NWKYGI-TFTGYFVNEKTIKYI 137
R+H HG+ G NG ++ N WK G T + EK+ YI
Sbjct 122 RIHFHGVTQGFSPPLVEARYPKNRRLIKRNGMQIYNAPRWKNGFSTVSRIQSKEKSASYI 181
Query 138 TKYMLK 143
TKY+ K
Sbjct 182 TKYISK 187
> zma:100216595 TIDP3618; LOC100216595
Length=322
Score = 37.4 bits (85), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 31/121 (26%), Positives = 54/121 (45%), Gaps = 10/121 (8%)
Query 7 ACGDC--YECRKQKQR-QWMVRMSEENRQTPNAYFLTLTIDDKSYKQIKQKYN-LKDNND 62
+C C +ECRKQK WMV+ + P+ FL + + +++K N LK +
Sbjct 112 SCSSCLFFECRKQKDLYLWMVK----SPGGPSVKFLVNAV--HTMEELKLTGNHLKGSRP 165
Query 63 IATKAIRLCLERVRKLTGKSVKHWFITELGHEKTERLHLHGIVWGLGNGEKVTNNWKYGI 122
+ T + + KL + + H F T H K + H H V+ + +G N++ +
Sbjct 166 LLTFSTNFDEQPHWKLVKEIITHIFATPKDHRKAKPFHDHVFVFSIVDGHVWFRNYQISV 225
Query 123 T 123
Sbjct 226 P 226
> dosa:Os12t0502300-01 Os12g0502300; Similar to Cyclin A-like
protein (Fragment).
Length=490
Score = 36.6 bits (83), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 38/154 (25%), Positives = 65/154 (42%), Gaps = 21/154 (14%)
Query 22 WMVRMSEENRQTPNAYFLTLTIDDKSYKQI---KQKYNLKDNND--IATKAIRLCLERVR 76
W+V +SEE + P+ +LT+ + D+ Q +QK L IA+K +C RV
Sbjct 267 WLVEVSEEYKLVPDTLYLTINLIDRFLSQHYIERQKLQLLGITSMLIASKYEEICAPRVE 326
Query 77 KLTGKSVKHWFITELGHEKTERLHLHGIVWG-LGNGEKVTNNWKYGITF-----TGYFVN 130
+ FIT+ + K E L + G+V +G V + F V
Sbjct 327 EF-------CFITDNTYTKAEVLKMEGLVLNDMGFHLSVPTTKTFLRRFLRAAQASRNVP 379
Query 131 EKTIKYITKYMLK---VDEKHPKFRGKVLCSAGI 161
T+ Y+ Y+ + +D KF V+ ++ +
Sbjct 380 SITLGYLANYLAELTLIDYSFLKFLPSVVAASAV 413
> osa:4352280 Os12g0502300
Length=490
Score = 36.6 bits (83), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 38/154 (25%), Positives = 65/154 (42%), Gaps = 21/154 (14%)
Query 22 WMVRMSEENRQTPNAYFLTLTIDDKSYKQI---KQKYNLKDNND--IATKAIRLCLERVR 76
W+V +SEE + P+ +LT+ + D+ Q +QK L IA+K +C RV
Sbjct 267 WLVEVSEEYKLVPDTLYLTINLIDRFLSQHYIERQKLQLLGITSMLIASKYEEICAPRVE 326
Query 77 KLTGKSVKHWFITELGHEKTERLHLHGIVWG-LGNGEKVTNNWKYGITF-----TGYFVN 130
+ FIT+ + K E L + G+V +G V + F V
Sbjct 327 EF-------CFITDNTYTKAEVLKMEGLVLNDMGFHLSVPTTKTFLRRFLRAAQASRNVP 379
Query 131 EKTIKYITKYMLK---VDEKHPKFRGKVLCSAGI 161
T+ Y+ Y+ + +D KF V+ ++ +
Sbjct 380 SITLGYLANYLAELTLIDYSFLKFLPSVVAASAV 413
Lambda K H a alpha
0.318 0.136 0.408 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 546803287272