bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-10_CDS_annotation_glimmer3.pl_2_4
Length=572
Score E
Sequences producing significant alignments: (Bits) Value
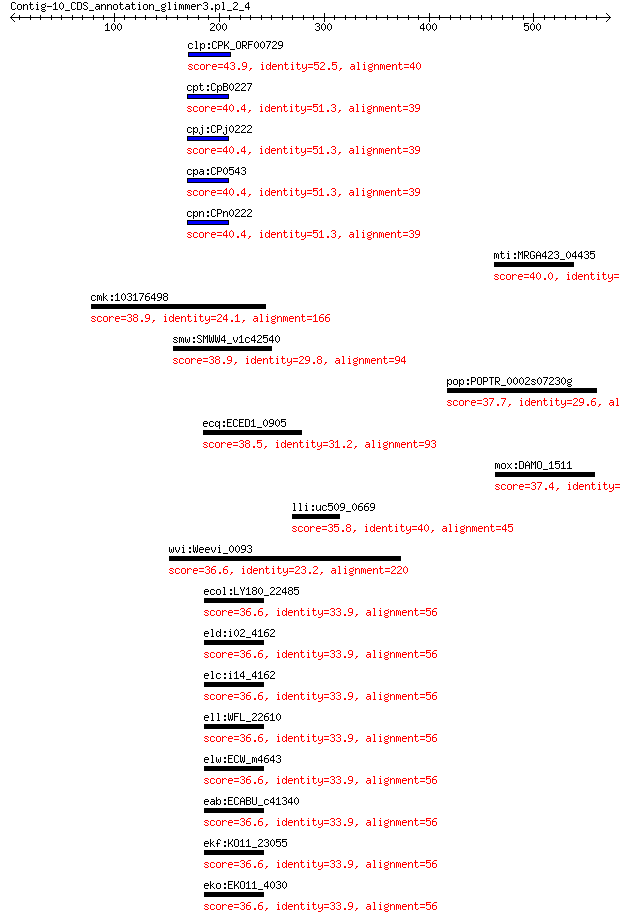
clp:CPK_ORF00729 hypothetical protein 43.9 0.013
cpt:CpB0227 hypothetical protein 40.4 0.17
cpj:CPj0222 hypothetical protein 40.4 0.17
cpa:CP0543 hypothetical protein 40.4 0.17
cpn:CPn0222 hypothetical protein 40.4 0.17
mti:MRGA423_04435 hypothetical protein 40.0 1.2
cmk:103176498 plekha3; pleckstrin homology domain containing, ... 38.9 2.4
smw:SMWW4_v1c42540 phage replication protein 38.9 3.4
pop:POPTR_0002s07230g POPTRDRAFT_754484; hypothetical protein 37.7 4.3
ecq:ECED1_0905 putative endonuclease from prophage, replicatio... 38.5 5.2
mox:DAMO_1511 Alpha/beta hydrolase fold 37.4 7.9
lli:uc509_0669 spxA; Arsenate reductase family protein 35.8 8.3
wvi:Weevi_0093 thymidylate synthase 36.6 8.9
ecol:LY180_22485 malate transporter 36.6 9.3
eld:i02_4162 hypothetical protein 36.6 9.3
elc:i14_4162 hypothetical protein 36.6 9.3
ell:WFL_22610 hypothetical protein 36.6 9.3
elw:ECW_m4643 hypothetical protein 36.6 9.3
eab:ECABU_c41340 hypothetical protein 36.6 9.3
ekf:KO11_23055 hypothetical protein 36.6 9.3
eko:EKO11_4030 hypothetical protein 36.6 9.3
> clp:CPK_ORF00729 hypothetical protein
Length=121
Score = 43.9 bits (102), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 21/40 (53%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 171 LDLFLKRLRSYYLDEKLRYYAVSEYGPTSFRPHWHLLLFS 210
L LFLKRLR K+RY+ EYG RPH+HLL+F+
Sbjct 75 LQLFLKRLRDRISPHKIRYFGCGEYGTKLQRPHYHLLIFN 114
> cpt:CpB0227 hypothetical protein
Length=113
Score = 40.4 bits (93), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 23/39 (59%), Gaps = 0/39 (0%)
Query 170 DLDLFLKRLRSYYLDEKLRYYAVSEYGPTSFRPHWHLLL 208
L LFLKRLR K+RY+ YG RPH+HLLL
Sbjct 74 HLQLFLKRLRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpj:CPj0222 hypothetical protein
Length=113
Score = 40.4 bits (93), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 23/39 (59%), Gaps = 0/39 (0%)
Query 170 DLDLFLKRLRSYYLDEKLRYYAVSEYGPTSFRPHWHLLL 208
L LFLKRLR K+RY+ YG RPH+HLLL
Sbjct 74 HLQLFLKRLRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpa:CP0543 hypothetical protein
Length=113
Score = 40.4 bits (93), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 23/39 (59%), Gaps = 0/39 (0%)
Query 170 DLDLFLKRLRSYYLDEKLRYYAVSEYGPTSFRPHWHLLL 208
L LFLKRLR K+RY+ YG RPH+HLLL
Sbjct 74 HLQLFLKRLRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpn:CPn0222 hypothetical protein
Length=113
Score = 40.4 bits (93), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 23/39 (59%), Gaps = 0/39 (0%)
Query 170 DLDLFLKRLRSYYLDEKLRYYAVSEYGPTSFRPHWHLLL 208
L LFLKRLR K+RY+ YG RPH+HLLL
Sbjct 74 HLQLFLKRLRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> mti:MRGA423_04435 hypothetical protein
Length=377
Score = 40.0 bits (92), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 38/77 (49%), Gaps = 12/77 (16%)
Query 462 PGGELFGRDRFLRIISEKIVTFWNRYDYNRL-VDFYQTLEDANDRDLVDFELRNYSFRYN 520
PGG++ G+ +L R+D RL VD ++ L + R +D LR RY+
Sbjct 195 PGGQVSGQRVWL-----------ARHDRKRLGVDHHRVLSTPSHRSTLDGLLRTGQARYS 243
Query 521 RPPDNKEKPYNKLPLVR 537
R PD++ P +L VR
Sbjct 244 RRPDDQPDPQGRLAPVR 260
> cmk:103176498 plekha3; pleckstrin homology domain containing,
family A (phosphoinositide binding specific) member 3
Length=316
Score = 38.9 bits (89), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 40/169 (24%), Positives = 71/169 (42%), Gaps = 16/169 (9%)
Query 78 VSIIETCSDDIADVPCVPNINE---LDDSDPNTYLFGFRNIPRSASIKLKNSTVERTFKD 134
+ + T D P N+NE L + NT++ + A+ K K + + D
Sbjct 134 IQQVHTMQDQPNLSPNAENMNEASSLLSATCNTFITTLEECMKIANAKFKPEMFQLSHSD 193
Query 135 PEVKFSYPMKPKELLSILGKVKHDVPNRIPYICNRDLDLFLKRLRSYYLDEKLRYYAVSE 194
P V P P +++ + + H P IPY +++ + + E + A +E
Sbjct 194 PLVSPVSP-SPVQMVRMKRSISH--PGTIPYD---------RKIETNHQKENIDVLARAE 241
Query 195 YGPTSFRPHWHLLLFSNSERFSETICENISKAWSYGRCDASLSRGFAAP 243
TSFR H + +S++E +S++ EN K R S++ G P
Sbjct 242 E-ITSFRSHRRIRTYSDTESYSDSALENAEKTAQNMRNLLSIANGEFTP 289
> smw:SMWW4_v1c42540 phage replication protein
Length=738
Score = 38.9 bits (89), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 28/105 (27%), Positives = 46/105 (44%), Gaps = 11/105 (10%)
Query 156 KHDVPNRIPYICNRDLDLFLKRLRSYYLDEKLRYYAVSEYGPT-SFRPHWHLLLFSNSER 214
K D P R L ++R+ + D L+ Y V P PHWH++LF++ E+
Sbjct 335 KWDEEAYTPKDGQRYLVKLFSKIRTAFKDAGLQVYGVRVVEPHHDATPHWHMMLFTSKEQ 394
Query 215 FSETI---------CENISKAWSYGRCD-ASLSRGFAAPYVASYV 249
+ I + + + R D L++G AA Y+A Y+
Sbjct 395 RQQVIDIMRRYAMAEDGDERGAAKNRFDCKHLNKGGAAGYIAKYI 439
> pop:POPTR_0002s07230g POPTRDRAFT_754484; hypothetical protein
Length=234
Score = 37.7 bits (86), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 42/159 (26%), Positives = 62/159 (39%), Gaps = 35/159 (22%)
Query 418 LYDGTDLESTHRLSRVYRFFLGISK-------FIRTYSVDG-----CSELFWSSGTPGGE 465
L + TD+E T L +V + G+ + + VDG C + SS
Sbjct 45 LLEATDIE-TQLLDKVLMYVRGMGEPNSLKEWVVMRLQVDGYEASLCKTSWVSS------ 97
Query 466 LFGRDRFLRIISEKIVTFWNRYDYNRLVDFYQTLEDANDRDLVDFELRNYSFRYNRPPDN 525
FG K++ F YDY ++ Q L + R +VD +LR+ F RP
Sbjct 98 -FGH---------KVIQFTGDYDYIDVMIMDQNLSNKTTRLIVDMDLRS-QFELARPTQT 146
Query 526 KEKPYNKLPLV-----RRLAAVALMKCRGKVKHKKLNDL 559
++ N LP V RL + + C K NDL
Sbjct 147 YKELINALPSVFVGSEERLDKIISLLCSAAKASLKENDL 185
> ecq:ECED1_0905 putative endonuclease from prophage, replication
protein A (GpA)
Length=840
Score = 38.5 bits (88), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 29/109 (27%), Positives = 46/109 (42%), Gaps = 18/109 (17%)
Query 185 EKLRYYAVSEYGPT-SFRPHWHLLLFS---NSERFSE------------TICENISKAWS 228
E L +Y V P PHWH+L+F+ N ER E + +IS +
Sbjct 433 EGLSWYGVRTVEPHHDGTPHWHMLVFTAPDNEERIIEIMRDAAIKSDRAELGNDISPRFK 492
Query 229 YGRCDASLSRGFAAPYVASYVNSFVALPNFYTQMPKVVRPKSFHSIGFT 277
+ D + +G A Y+A+Y+ + F+ PK +P + G T
Sbjct 493 CEKIDPA--KGTPASYIATYIGKNLDASTFHDNDPKTGKPYADKESGKT 539
> mox:DAMO_1511 Alpha/beta hydrolase fold
Length=356
Score = 37.4 bits (85), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 36/111 (32%), Positives = 51/111 (46%), Gaps = 16/111 (14%)
Query 463 GGELFGRDRFL----RIISEKIVTFWNRYD------YNRLVDFYQTLEDANDRDLVDFEL 512
G +LF R++ L +I+ E ++ F + ++ + Y LE +R F L
Sbjct 47 GVKLFVREKRLANLPKIVKENVILFVHGVPGPGSVMFDLAIPGYSWLEYVAERGFDAFTL 106
Query 513 RNYSF-RYNRPPDNKEKPYNKLPLVR-----RLAAVALMKCRGKVKHKKLN 557
F R RPP+ KE PY LPLVR R VA+ R K K K+N
Sbjct 107 DIRGFGRSTRPPEMKESPYQNLPLVRADQVMRDIDVAVDYIRSKRKVDKVN 157
> lli:uc509_0669 spxA; Arsenate reductase family protein
Length=132
Score = 35.8 bits (81), Expect = 8.3, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 270 SFHSIGFTESNLFPRKVRITEIDEVTNKCLDGVSVERNGRFRTIK 314
S+H I F E NL + TEI ++ KC DGV + R R +K
Sbjct 21 SYHHIPFNERNLIADPLSTTEISQILQKCDDGVEGLISSRNRFVK 65
> wvi:Weevi_0093 thymidylate synthase
Length=211
Score = 36.6 bits (83), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 51/223 (23%), Positives = 88/223 (39%), Gaps = 43/223 (19%)
Query 153 GKVKHDVPNRIPYICNRDLDLFLKRLRSYYLDEKLRYYAVSEYGPTSFRPHWHLLLFSNS 212
GKV+ + I Y+ N+ L+L L + E+G + L LF+
Sbjct 15 GKVQSNKKGNITYLLNQKLELKPTDLLEIF----------EEHGIARNKLKTELELFTKG 64
Query 213 ERFSETICENISKAWSYGRCDASLSRGFAAPYVASYVNSFVALPNFYTQMPKVVRPKSFH 272
ER +E EN + W Y C L VNS+ P ++ Q+P +++ +
Sbjct 65 ERLTERYRENGIEWWDY--CGPIL------------VNSY---PTYFEQLPDLIKKINKE 107
Query 273 SIGFTESNLFPRKVRITEIDEVTNKCLDGVSVE-RNGRFRTIKPTWPYLLRLFPRFSDPI 331
LF + T+++ CL V + G+ ++ + R SD
Sbjct 108 KRNSKNYVLFLGR---TDVESNQQPCLSLVQFQIEKGKL---------IISAYQRSSDAS 155
Query 332 RKLPSSVYQLLLSA--LSAPARVIRGGCADITCDPFNVNAPSK 372
LP+ +Y L L + ++ P + I A++ NV P+K
Sbjct 156 LGLPADIYHLYLISRQINLPLKSITLMLANVHIYENNV-EPTK 197
> ecol:LY180_22485 malate transporter
Length=204
Score = 36.6 bits (83), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 19/56 (34%), Positives = 28/56 (50%), Gaps = 0/56 (0%)
Query 186 KLRYYAVSEYGPTSFRPHWHLLLFSNSERFSETICENISKAWSYGRCDASLSRGFA 241
KLRY V E+G + H+H+LL N E F T+ + A G ++R +A
Sbjct 84 KLRYAWVREFGELNGNKHYHMLLLVNREVFRNTMLNTENSAQPRGLLVGMVARAWA 139
> eld:i02_4162 hypothetical protein
Length=204
Score = 36.6 bits (83), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 19/56 (34%), Positives = 28/56 (50%), Gaps = 0/56 (0%)
Query 186 KLRYYAVSEYGPTSFRPHWHLLLFSNSERFSETICENISKAWSYGRCDASLSRGFA 241
KLRY V E+G + H+H+LL N E F T+ + A G ++R +A
Sbjct 84 KLRYAWVREFGELNGNKHYHMLLLVNREVFRNTMLNTENSAQPRGLLVGMVARAWA 139
> elc:i14_4162 hypothetical protein
Length=204
Score = 36.6 bits (83), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 19/56 (34%), Positives = 28/56 (50%), Gaps = 0/56 (0%)
Query 186 KLRYYAVSEYGPTSFRPHWHLLLFSNSERFSETICENISKAWSYGRCDASLSRGFA 241
KLRY V E+G + H+H+LL N E F T+ + A G ++R +A
Sbjct 84 KLRYAWVREFGELNGNKHYHMLLLVNREVFRNTMLNTENSAQPRGLLVGMVARAWA 139
> ell:WFL_22610 hypothetical protein
Length=204
Score = 36.6 bits (83), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 19/56 (34%), Positives = 28/56 (50%), Gaps = 0/56 (0%)
Query 186 KLRYYAVSEYGPTSFRPHWHLLLFSNSERFSETICENISKAWSYGRCDASLSRGFA 241
KLRY V E+G + H+H+LL N E F T+ + A G ++R +A
Sbjct 84 KLRYAWVREFGELNGNKHYHMLLLVNREVFRNTMLNTENSAQPRGLLVGMVARAWA 139
> elw:ECW_m4643 hypothetical protein
Length=204
Score = 36.6 bits (83), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 19/56 (34%), Positives = 28/56 (50%), Gaps = 0/56 (0%)
Query 186 KLRYYAVSEYGPTSFRPHWHLLLFSNSERFSETICENISKAWSYGRCDASLSRGFA 241
KLRY V E+G + H+H+LL N E F T+ + A G ++R +A
Sbjct 84 KLRYAWVREFGELNGNKHYHMLLLVNREVFRNTMLNTENSAQPRGLLVGMVARAWA 139
> eab:ECABU_c41340 hypothetical protein
Length=204
Score = 36.6 bits (83), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 19/56 (34%), Positives = 28/56 (50%), Gaps = 0/56 (0%)
Query 186 KLRYYAVSEYGPTSFRPHWHLLLFSNSERFSETICENISKAWSYGRCDASLSRGFA 241
KLRY V E+G + H+H+LL N E F T+ + A G ++R +A
Sbjct 84 KLRYAWVREFGELNGNKHYHMLLLVNREVFRNTMLNTENSAQPRGLLVGMVARAWA 139
> ekf:KO11_23055 hypothetical protein
Length=204
Score = 36.6 bits (83), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 19/56 (34%), Positives = 28/56 (50%), Gaps = 0/56 (0%)
Query 186 KLRYYAVSEYGPTSFRPHWHLLLFSNSERFSETICENISKAWSYGRCDASLSRGFA 241
KLRY V E+G + H+H+LL N E F T+ + A G ++R +A
Sbjct 84 KLRYAWVREFGELNGNKHYHMLLLVNREVFRNTMLNTENSAQPRGLLVGMVARAWA 139
> eko:EKO11_4030 hypothetical protein
Length=204
Score = 36.6 bits (83), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 19/56 (34%), Positives = 28/56 (50%), Gaps = 0/56 (0%)
Query 186 KLRYYAVSEYGPTSFRPHWHLLLFSNSERFSETICENISKAWSYGRCDASLSRGFA 241
KLRY V E+G + H+H+LL N E F T+ + A G ++R +A
Sbjct 84 KLRYAWVREFGELNGNKHYHMLLLVNREVFRNTMLNTENSAQPRGLLVGMVARAWA 139
Lambda K H a alpha
0.323 0.139 0.431 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1305383765826