bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-10_CDS_annotation_glimmer3.pl_2_1
Length=279
Score E
Sequences producing significant alignments: (Bits) Value
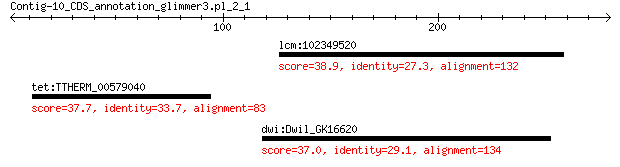
lcm:102349520 MRVI1; murine retrovirus integration site 1 homolog 38.9 1.3
tet:TTHERM_00579040 cation channel family protein 37.7 2.7
dwi:Dwil_GK16620 GK16620 gene product from transcript GK16620-RA 37.0 5.3
> lcm:102349520 MRVI1; murine retrovirus integration site 1 homolog
Length=1139
Score = 38.9 bits (89), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 36/133 (27%), Positives = 59/133 (44%), Gaps = 14/133 (11%)
Query 126 LDNEAKGILNKYLDQEQQLDLNVKAADYYQRMSAGYLSYAETKKALADEALAAARTRGQK 185
L A + K Q ++D V A + LS AE+K+ALA+E L AA R ++
Sbjct 181 LKESASETVAKSYKQVTEMDDKVSKATLMMHAAQEKLSVAESKRALAEEKLKAAMERVEE 240
Query 186 ISNEVASRIAESQIAANIAANQSSEAYHNEELKLGLSQDNA-RSKNIEEWYRSRNEKKRY 244
+ ++ + +A AA + S + LG+SQ RS I E + E+++
Sbjct 241 VQCQLVA------LAERTAATEQS-------MDLGMSQAACTRSSKIRELHFQTKEEEKL 287
Query 245 KYYDADKWVEYGT 257
+ D V+Y
Sbjct 288 VSFTVDHNVQYCC 300
> tet:TTHERM_00579040 cation channel family protein
Length=1469
Score = 37.7 bits (86), Expect = 2.7, Method: Composition-based stats.
Identities = 28/83 (34%), Positives = 38/83 (46%), Gaps = 6/83 (7%)
Query 11 QSAGNGATAASSGNAVMQPFQADYSGIGSSIGNIFQYDLMQSEKSQLQGARQLSDAKAME 70
Q+ GN T N P DYS G S NIFQYD + Q A L+ A+A++
Sbjct 180 QTLGNNNTF----NIWDNPITPDYSTYGPS--NIFQYDSYNNGFVQFFSAFNLTHAQAVD 233
Query 71 TLSNIDWGNLTAETRNYLRSTGL 93
L+N+ E NY+ G+
Sbjct 234 MLTNLSNDQFFDEQTNYISCDGV 256
> dwi:Dwil_GK16620 GK16620 gene product from transcript GK16620-RA
Length=1645
Score = 37.0 bits (84), Expect = 5.3, Method: Composition-based stats.
Identities = 39/139 (28%), Positives = 59/139 (42%), Gaps = 9/139 (6%)
Query 118 RAQRSGMLLDNEAKGILNKYLDQEQQLDLNVKAADYYQRMSAGYLSYAETKKALADEALA 177
+ ++ + L AK K +Q D +K A+ AG+ S E K AD+AL
Sbjct 1366 KQKQDDIELLERAKAAFEKATKAVKQGDNTLKEANNTYNTLAGFQSDVEASKEKADQALQ 1425
Query 178 AARTRGQKISN--EVASRIAESQIAANIAANQSSEAYHNEELKLGLSQDNARSKNIEEWY 235
+ Q+I N + S+ E+ AN AN++ E + K SK+ E
Sbjct 1426 TVPSIEQEIQNAKHLISQADEALDGANKNANEAKENAQEAQKKYA----EQASKDAELIR 1481
Query 236 RSRNEKK---RYKYYDADK 251
R NE K R Y+AD+
Sbjct 1482 RKANETKVAARNLRYEADQ 1500
Lambda K H a alpha
0.311 0.126 0.343 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 445107121113