bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggnogv4.proteins.all.fa
14,875,530 sequences; 5,112,597,290 total letters
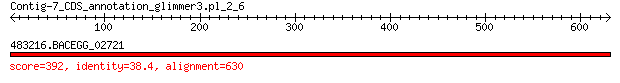
Query= Contig-7_CDS_annotation_glimmer3.pl_2_6
Length=630
Score E
Sequences producing significant alignments: (Bits) Value
483216.BACEGG_02721 392 3e-125
> 483216.BACEGG_02721
Length=578
Score = 392 bits (1008), Expect = 3e-125, Method: Compositional matrix adjust.
Identities = 242/648 (37%), Positives = 352/648 (54%), Gaps = 88/648 (14%)
Query 1 MAHFTGLKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRP 60
MA+ LK ++N P R+GFD+ K F+AK GELLPV +PG T+ I+++ FTRT+P
Sbjct 1 MANIMSLKSIRNKPSRNGFDLSFKKNFTAKAGELLPVMVKEVLPGDTFKINLKAFTRTQP 60
Query 61 VQTAAYTRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDI--LTALTVSGDLPYC 118
V TAA+ RIREY+DF+ VP DL+W + + QM + P A I +SG++PY
Sbjct 61 VNTAAFARIREYYDFFFVPYDLLWNKANTVLTQMYDN-PQHAVSIDPTRNFVLSGEMPYM 119
Query 119 SLSDLGLSCFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGN---IIPN 175
+ + S + ++ KS N FGY R + KL+ L YGN + +
Sbjct 120 TSEAIASYINALSTASALADYKS--------NYFGYNRSKSSVKLLEYLGYGNYESFLTD 171
Query 176 NMPALNIGNSNYRWWNREAPLASDSLIVYSQMYNFNMNVNLFPLATYQKIYQDFFRWSQW 235
+ WN APL + N+N N+F L YQKIY DF+R SQW
Sbjct 172 D-------------WN-TAPLMA------------NLNHNIFGLLAYQKIYSDFYRDSQW 205
Query 236 EKADPTSYNFDWYQGSGNLFGGTIDTSLPASSDYWKRDNLFSLRYCNWNKDLFMGVLPNS 295
E+ P+++N D+ GS +++ S+++++ N F LRYCNW KDLF GVLP+
Sbjct 206 ERVSPSTFNVDYLDGS------SMNLDNAYSTEFYQNYNFFDLRYCNWQKDLFHGVLPHQ 259
Query 296 QFGDIAVIDIEGGLNIPASRISLSSNNRPTIGIKVGAQVSSPNNCSITNSSGNLSTGDIL 355
Q+G+ AV I P L+ +N T+G +SP S T ++ NL D
Sbjct 260 QYGETAVASI-----TPDVTGKLTLSNFSTVG-------TSPTTASGT-ATKNLPAFD-- 304
Query 356 SVGIPAASYKLQSSFNVLALRQAESLQKYREITQSVDTNYRDQIKAHFGVNVPASDSHMA 415
+VG ++L LRQAE LQK++EITQS + +Y+DQ++ H+GV+V S +
Sbjct 305 TVG----------DLSILVLRQAEFLQKWKEITQSGNKDYKDQLEKHWGVSVGDGFSELC 354
Query 416 QYIGGIARNLDISEVVNNNLQGDGEAVIYGKGVGTGTGSMRYTTGSKYCILMCIYHCMPV 475
Y+GG++ ++DI+EV+N N+ G A I GKGVG G + + + +Y ++MCIYHC+P+
Sbjct 355 TYLGGVSSSIDINEVINTNITGSAAADIAGKGVGVANGEINFNSNGRYGLIMCIYHCLPL 414
Query 476 LDYDISGQHPQLLATSVDELPIPEFDNIGMEGVPLVQLVNSNLYKTNKSVKIDSILGYNP 535
LDY P L + + IPEFD +GM+ +PLVQL+N N S +LGY P
Sbjct 415 LDYTTDMLDPAFLKVNSTDYAIPEFDRVGMQSMPLVQLMNPLRSFANAS---GLVLGYVP 471
Query 536 RYYAWKSNIDRIHGAFTTTLQDWV-------------SPVDDSFLYSTFGTPSSGSFVTW 582
RY +K+++D+ G F TL WV P D + + PS + +
Sbjct 472 RYIDYKTSVDQSVGGFKRTLNSWVISYGNISVLKQVTLPNDAPPIEPSEPVPSVAP-MNF 530
Query 583 PFFKVNPNTLDNIFAVKSDSTWESDQFLVNSYVGCKVVRPLSRDGVPY 630
FFKVNP+ LD IFAV++ +DQFL +S+ K VR L DG+PY
Sbjct 531 TFFKVNPDCLDPIFAVQAGDDTNTDQFLCSSFFDIKAVRNLDTDGLPY 578
Lambda K H a alpha
0.319 0.136 0.422 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1423332636440