bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggnogv4.proteins.all.fa
14,875,530 sequences; 5,112,597,290 total letters
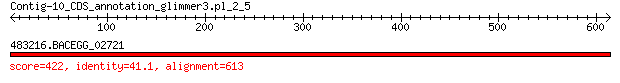
Query= Contig-10_CDS_annotation_glimmer3.pl_2_5
Length=613
Score E
Sequences producing significant alignments: (Bits) Value
483216.BACEGG_02721 422 5e-137
> 483216.BACEGG_02721
Length=578
Score = 422 bits (1085), Expect = 5e-137, Method: Compositional matrix adjust.
Identities = 252/622 (41%), Positives = 345/622 (55%), Gaps = 53/622 (9%)
Query 1 MASYTGMSNLQNHPHRSGFDIGRKNAFTAKVGELLPVYWDISMPGDKYRFNIEYFTRTQP 60
MA+ + +++N P R+GFD+ K FTAK GELLPV +PGD ++ N++ FTRTQP
Sbjct 1 MANIMSLKSIRNKPSRNGFDLSFKKNFTAKAGELLPVMVKEVLPGDTFKINLKAFTRTQP 60
Query 61 VETSAYTRLREYFDFYAVPLRLLWKSAPSVLTQMQDVNKIQALSL--TQNLSLGTFLPSI 118
V T+A+ R+REY+DF+ VP LLW A +VLTQM D N A+S+ T+N L +P +
Sbjct 61 VNTAAFARIREYYDFFFVPYDLLWNKANTVLTQMYD-NPQHAVSIDPTRNFVLSGEMPYM 119
Query 119 PLSVLSDAMYLLNGRSWTPGNSSSLKNMFGFDRSDLCYKLLSYLGYGNLISSESTGRWWS 178
++ +N S + N FG++RS KLL YLGYGN S T W
Sbjct 120 TSEAIAS---YINALSTASALADYKSNYFGYNRSKSSVKLLEYLGYGNY-ESFLTDDW-- 173
Query 179 TSVSSKNDASYTQRYIQNNYVNLFPLLAYQKIYQDFFRWSQWEASNPSSYNVDYFTGVSP 238
T + N N+F LLAYQKIY DF+R SQWE +PS++NVDY G S
Sbjct 174 ----------NTAPLMANLNHNIFGLLAYQKIYSDFYRDSQWERVSPSTFNVDYLDGSSM 223
Query 239 SLISSLLSVSPDYWKSGTMFDLKYCNWNKDMLMGVLPNSQFGDVAVLDIPDSGDSNVVLG 298
+L ++ S +++++ FDL+YCNW KD+ GVLP+ Q+G+ AV I + L
Sbjct 224 NLDNAY---STEFYQNYNFFDLRYCNWQKDLFHGVLPHQQYGETAVASITPDVTGKLTL- 279
Query 299 TDSHKSSVGIASAITSKTAPFPLFALDASPENPIPINSKLRLDLSSLKSQFTVLALRQAE 358
S+ S+VG + S TA L A D + ++L LRQAE
Sbjct 280 --SNFSTVGTSPTTASGTATKNLPAFDTVGD-------------------LSILVLRQAE 318
Query 359 ALQRWKEISQSGDSDYREQIRKHFGVKLPQALSNMCTYIGGVSRNLDISEVVNNNLATEG 418
LQ+WKEI+QSG+ DY++Q+ KH+GV + S +CTY+GGVS ++DI+EV+N N+ T
Sbjct 319 FLQKWKEITQSGNKDYKDQLEKHWGVSVGDGFSELCTYLGGVSSSIDINEVINTNI-TGS 377
Query 419 DTAVIAGKGVGAGNGSFEYTTT-EHCVVMCIYHAVPLLDYTLTGQDGQLLVTDAESLPIP 477
A IAGKGVG NG + + + ++MCIYH +PLLDYT D L ++ IP
Sbjct 378 AAADIAGKGVGVANGEINFNSNGRYGLIMCIYHCLPLLDYTTDMLDPAFLKVNSTDYAIP 437
Query 478 EFDNIGMEVLPMTQVFNSPKASIVNL--FNAGYNPRYFNWKTKLDVINGAFTTTLKSWVS 535
EFD +GM+ +P+ Q+ N P S N GY PRY ++KT +D G F TL SWV
Sbjct 438 EFDRVGMQSMPLVQLMN-PLRSFANASGLVLGYVPRYIDYKTSVDQSVGGFKRTLNSWVI 496
Query 536 PVTESLLSGWFCFGYNKDDAAPDTKV----IMNYKFFKVNPSVLDPIFGVNADSTWDTDQ 591
+ + P V MN+ FFKVNP LDPIF V A +TDQ
Sbjct 497 SYGNISVLKQVTLPNDAPPIEPSEPVPSVAPMNFTFFKVNPDCLDPIFAVQAGDDTNTDQ 556
Query 592 LLVNSYIGCYVVRNLSRDGVPY 613
L +S+ VRNL DG+PY
Sbjct 557 FLCSSFFDIKAVRNLDTDGLPY 578
Lambda K H a alpha
0.318 0.134 0.408 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1373339547970