bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-9_CDS_annotation_glimmer3.pl_2_7
Length=157
Score E
Sequences producing significant alignments: (Bits) Value
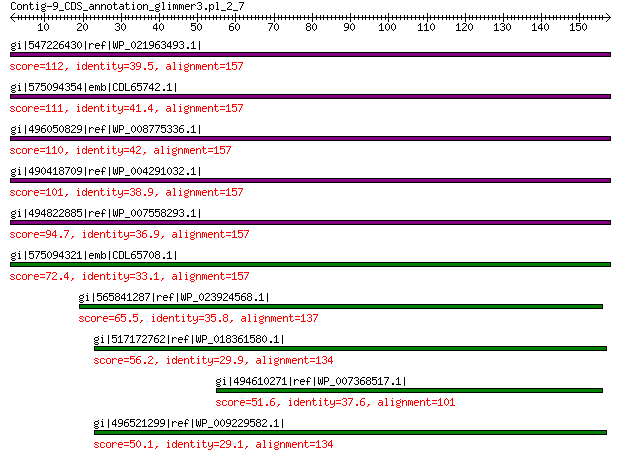
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 112 6e-26
gi|575094354|emb|CDL65742.1| unnamed protein product 111 2e-25
gi|496050829|ref|WP_008775336.1| hypothetical protein 110 2e-25
gi|490418709|ref|WP_004291032.1| hypothetical protein 101 4e-22
gi|494822885|ref|WP_007558293.1| hypothetical protein 94.7 1e-19
gi|575094321|emb|CDL65708.1| unnamed protein product 72.4 8e-12
gi|565841287|ref|WP_023924568.1| hypothetical protein 65.5 1e-09
gi|517172762|ref|WP_018361580.1| hypothetical protein 56.2 2e-06
gi|494610271|ref|WP_007368517.1| capsid protein 51.6 5e-05
gi|496521299|ref|WP_009229582.1| capsid protein 50.1 2e-04
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 112 bits (280), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 62/161 (39%), Positives = 95/161 (59%), Gaps = 8/161 (5%)
Query 1 LDYQLTGPDLQLLNTYATDLPQPELDNLGLEAL--PYFTFVNDAVATQPNNVTVKSIIGY 58
LD+ + Q T TD PE D++G++ L F + + + P+++ +GY
Sbjct 417 LDWSINRIARQNFKTTFTDYAIPEFDSVGMQQLYPSEMIFGLEDLPSDPSSIN----MGY 472
Query 59 VPRYIAYKTDIDCVDGAFLTSLTSWVTPLTIDEIVT--KISLGSGTGPFTPNYGLFKVSP 116
VPRY KT ID + G+F+ +L SWV+PLT I + +G T Y FKV+P
Sbjct 473 VPRYADLKTSIDEIHGSFIDTLVSWVSPLTDSYISAYRQACKDAGFSDITMTYNFFKVNP 532
Query 117 YVLDSIFVSQCDSTVDTDQFLVESFFDVKLVQNLDYNGMPY 157
+++D+IF + DST++TDQ L+ S+FD+K V+N DYNG+PY
Sbjct 533 HIVDNIFGVKADSTINTDQLLINSYFDIKAVRNFDYNGLPY 573
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 111 bits (277), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 65/164 (40%), Positives = 91/164 (55%), Gaps = 12/164 (7%)
Query 1 LDYQLTGPDLQLLNTYATDLPQPELDNLGLEALPYFTFVNDAVATQPNNVTVKSIIGYVP 60
+DY +G D AT P PELD +G+E++P +N + + + + +GY P
Sbjct 457 VDYVGSGVDHSCTLVDATSFPIPELDQIGMESVPLVRAMNPV--KESDTPSADTFLGYAP 514
Query 61 RYIAYKTDIDCVDGAFLTSLTSWVTPLTIDEIVTKISLGSGTGPFTPN-------YGLFK 113
RYI +KT +D G F SL +W P+ E+ + SL P PN G FK
Sbjct 515 RYIDWKTSVDRSVGDFADSLRTWCLPVGDKELTSANSLNF---PSNPNVEPDSIAAGFFK 571
Query 114 VSPYVLDSIFVSQCDSTVDTDQFLVESFFDVKLVQNLDYNGMPY 157
V+P ++D +F DSTV TD+FL SFFDVK+V+NLD NG+PY
Sbjct 572 VNPSIVDPLFAVVADSTVKTDEFLCSSFFDVKVVRNLDVNGLPY 615
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 110 bits (276), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 66/162 (41%), Positives = 94/162 (58%), Gaps = 10/162 (6%)
Query 1 LDY--QLTGPDLQLLNTYATDLPQPELDNLGLEALPYFTFVNDAVATQPNNVTVKSIIGY 58
LDY L P +N+ TD PE D +G+E++P + +N Q + SI+GY
Sbjct 424 LDYTTDLVNPAFTKINS--TDFAIPEFDRVGMESVPLVSLMN---PLQSSYNVGSSILGY 478
Query 59 VPRYIAYKTDIDCVDGAFLTSLTSWVTPLTIDEIVTKISLGS--GTGPFT-PNYGLFKVS 115
PRYI+YKTD+D GAF T+L SWV ++ +++ P T NY FKV+
Sbjct 479 APRYISYKTDVDSSVGAFKTTLKSWVMSYDNQSVINQLNYQDDPNNSPGTLVNYTNFKVN 538
Query 116 PYVLDSIFVSQCDSTVDTDQFLVESFFDVKLVQNLDYNGMPY 157
P +D +F +++DTDQFL SFFDVK+V+NLD +G+PY
Sbjct 539 PNCVDPLFAVAASNSIDTDQFLCSSFFDVKVVRNLDTDGLPY 580
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 101 bits (252), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 61/167 (37%), Positives = 88/167 (53%), Gaps = 13/167 (8%)
Query 1 LDYQLTGPDLQLLNTYATDLPQPELDNLGLEALPYFTFVNDAVATQPNNVTVKSIIGYVP 60
LDY D L +TD PE D +G++++P +N + + N + ++GYVP
Sbjct 415 LDYTTDMLDPAFLKVNSTDYAIPEFDRVGMQSMPLVQLMN-PLRSFANASGL--VLGYVP 471
Query 61 RYIAYKTDIDCVDGAFLTSLTSWVTPLTIDEIVTKISLGSGTGPFTP----------NYG 110
RYI YKT +D G F +L SWV ++ +++L + P P N+
Sbjct 472 RYIDYKTSVDQSVGGFKRTLNSWVISYGNISVLKQVTLPNDAPPIEPSEPVPSVAPMNFT 531
Query 111 LFKVSPYVLDSIFVSQCDSTVDTDQFLVESFFDVKLVQNLDYNGMPY 157
FKV+P LD IF Q +TDQFL SFFD+K V+NLD +G+PY
Sbjct 532 FFKVNPDCLDPIFAVQAGDDTNTDQFLCSSFFDIKAVRNLDTDGLPY 578
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 94.7 bits (234), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 58/165 (35%), Positives = 85/165 (52%), Gaps = 11/165 (7%)
Query 1 LDYQLTGPDLQLLNTYATDLPQPELDNLGLEALPYFTFVNDAVATQPN-NVTVKSIIGYV 59
LDY + P T D P PE D +G+E +P +N + V+ GY
Sbjct 452 LDYITSAPHFGTTLTNVLDFPIPEFDKIGMEQVPVIRGLNPVKPKDGDFKVSPNLYFGYA 511
Query 60 PRYIAYKTDIDCVDGAFLTSLTSWVTPLTIDEIVTKISLGSGTGPFTPN-------YGLF 112
P+Y +KT +D G F SL +W+ P + ++ S+ P PN G F
Sbjct 512 PQYYNWKTTLDKSMGEFRRSLKTWIIPFDDEALLAADSVDF---PDNPNVEADSVKAGFF 568
Query 113 KVSPYVLDSIFVSQCDSTVDTDQFLVESFFDVKLVQNLDYNGMPY 157
KVSP VLD++F + +S ++TDQFL + FDV +V++LD NG+PY
Sbjct 569 KVSPSVLDNLFAVKANSDLNTDQFLCSTLFDVNVVRSLDPNGLPY 613
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 72.4 bits (176), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 52/170 (31%), Positives = 75/170 (44%), Gaps = 20/170 (12%)
Query 1 LDYQLTGPDLQLLNTYATDLPQPELDNLGLEALPYFTFVNDAVATQPNNVTVKSI----- 55
LD+ G D L T A+D PE+D++G++ TF + A P N K+
Sbjct 478 LDFAHLGIDRTLFKTDASDFVIPEMDSIGMQQ----TFRCEVAAPAPYNDEFKAFRVGDG 533
Query 56 --------IGYVPRYIAYKTDIDCVDGAFLTSLTSWVTPLTIDEIVTKISLGSGTGPFTP 107
GY PRY +KT D +GAF SL SWVT + D I + + G P
Sbjct 534 SSPDMSETYGYAPRYSEFKTSYDRYNGAFCHSLKSWVTGINFDAIQNNV-WNTWAGINAP 592
Query 108 NYGLFKVSPYVLDSIFVSQCDSTVDTDQFLVESFFDVKLVQNLDYNGMPY 157
N +F P ++ ++F+ + D DQ V +NL G+PY
Sbjct 593 N--MFACRPDIVKNLFLVSSTNNSDDDQLYVGMVNMCYATRNLSRYGLPY 640
>gi|565841287|ref|WP_023924568.1| hypothetical protein [Prevotella nigrescens]
gi|564729907|gb|ETD29851.1| hypothetical protein HMPREF1173_00033 [Prevotella nigrescens
CC14M]
Length=656
Score = 65.5 bits (158), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 49/150 (33%), Positives = 73/150 (49%), Gaps = 33/150 (22%)
Query 19 DLPQPELDNLGLEALPYFTF---VNDA---VATQPNNVTVKSIIGYVPRYIAYKTDIDCV 72
D QPE +NLG++ + +N A + Q NNV +GY RY+ YKT D +
Sbjct 524 DYFQPEFENLGMQPVIQSDLCLCINSAKSDSSDQHNNV-----LGYSARYLEYKTARDII 578
Query 73 DGAFLT--SLTSWVTPLTIDEIVTKISLGSGTGPFTPNYGLFK-----VSPYVLDSIFVS 125
G F++ SL++W TP +T +G V P VL+ IF
Sbjct 579 FGEFMSGGSLSAWATP---------------KNNYTFEFGKLSLPDLLVDPKVLEPIFAV 623
Query 126 QCDSTVDTDQFLVESFFDVKLVQNLDYNGM 155
+ + ++ TDQFLV S+FDVK ++ + N M
Sbjct 624 KYNGSMSTDQFLVNSYFDVKAIRPMQVNDM 653
>gi|517172762|ref|WP_018361580.1| hypothetical protein [Prevotella nanceiensis]
Length=568
Score = 56.2 bits (134), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 40/142 (28%), Positives = 66/142 (46%), Gaps = 24/142 (17%)
Query 23 PELDNLGLEALPYFTFVNDAVATQPNNVTVKSII------GYVPRYIAYKTDIDCVDGAF 76
PE +NLG++ L F + ++ + NN T S I G+ PRY YKT +D G F
Sbjct 441 PEFENLGMQPL----FAKN-ISYKYNNNTANSRIKNLGAFGWQPRYSEYKTALDINHGQF 495
Query 77 LTS--LTSWVTPLTIDEIVTKISLGSGTGPFTPNYGLFKVSPYVLDSIFVSQCDSTVDTD 134
+ L+ W E ++ ++ + FK++P LD +F + T TD
Sbjct 496 VHQEPLSYWTVARARGESMSNFNIST-----------FKINPKWLDDVFAVNYNGTELTD 544
Query 135 QFLVESFFDVKLVQNLDYNGMP 156
Q +F++ V ++ +GMP
Sbjct 545 QVFGGCYFNIVKVSDMSIDGMP 566
>gi|494610271|ref|WP_007368517.1| capsid protein [Prevotella multiformis]
gi|324988543|gb|EGC20506.1| putative capsid protein (F protein) [Prevotella multiformis DSM
16608]
Length=531
Score = 51.6 bits (122), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 38/106 (36%), Positives = 56/106 (53%), Gaps = 12/106 (11%)
Query 55 IIGYVPRYIAYKTDIDCVDGAFLT--SLTSWVTP---LTIDEIVTKISLGSGTGPFTPNY 109
++G+ RY YKT D V G F + SL+ W +P D L P++P +
Sbjct 430 LLGWQVRYNEYKTSRDLVFGEFESGLSLSYWCSPRYDFGFDGKAGDKKLV--NSPWSPAH 487
Query 110 GLFKVSPYVLDSIFVSQCDSTVDTDQFLVESFFDVKLVQNLDYNGM 155
F V+P +L++IF+ S V D FLV SFFDVK V+ + +G+
Sbjct 488 --FYVNPSILNTIFLV---SAVKADHFLVNSFFDVKAVRPMSVSGL 528
>gi|496521299|ref|WP_009229582.1| capsid protein [Prevotella sp. oral taxon 317]
gi|288330570|gb|EFC69154.1| putative capsid protein (F protein) [Prevotella sp. oral taxon
317 str. F0108]
Length=541
Score = 50.1 bits (118), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 39/138 (28%), Positives = 60/138 (43%), Gaps = 24/138 (17%)
Query 23 PELDNLGLEAL-PYFTFVNDAVATQPNNVTVKSIIGYVPRYIAYKTDIDCVDGAFLTS-- 79
PE +NLG++ + P F +N A G+ PRY YKT D G F
Sbjct 422 PEFENLGMQPIVPAFVSLNRAKDNS---------YGWQPRYSEYKTAFDINHGQFANGEP 472
Query 80 LTSWVTPLTIDEIVTKISLGSGTGPF-TPNYGLFKVSPYVLDSIFVSQCDSTVDTDQFLV 138
L+ W I+ G+ T N K++P+ LDS+F + T TD
Sbjct 473 LSYW-----------SIARARGSDTLNTFNVAALKINPHWLDSVFAVNYNGTEVTDCMFG 521
Query 139 ESFFDVKLVQNLDYNGMP 156
+ F+++ V ++ +GMP
Sbjct 522 YAHFNIEKVSDMTEDGMP 539
Lambda K H a alpha
0.319 0.139 0.416 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 432232358643