bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-9_CDS_annotation_glimmer3.pl_2_3
Length=561
Score E
Sequences producing significant alignments: (Bits) Value
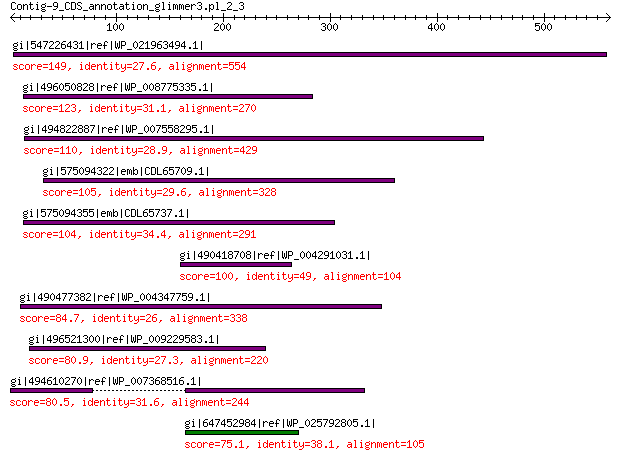
gi|547226431|ref|WP_021963494.1| predicted protein 149 1e-35
gi|496050828|ref|WP_008775335.1| hypothetical protein 123 7e-27
gi|494822887|ref|WP_007558295.1| hypothetical protein 110 3e-22
gi|575094322|emb|CDL65709.1| unnamed protein product 105 7e-21
gi|575094355|emb|CDL65737.1| unnamed protein product 104 2e-20
gi|490418708|ref|WP_004291031.1| hypothetical protein 100 1e-19
gi|490477382|ref|WP_004347759.1| hypothetical protein 84.7 4e-14
gi|496521300|ref|WP_009229583.1| hypothetical protein 80.9 8e-13
gi|494610270|ref|WP_007368516.1| hypothetical protein 80.5 8e-13
gi|647452984|ref|WP_025792805.1| hypothetical protein 75.1 5e-11
>gi|547226431|ref|WP_021963494.1| predicted protein [Prevotella sp. CAG:1185]
gi|524103383|emb|CCY83995.1| predicted protein [Prevotella sp. CAG:1185]
Length=498
Score = 149 bits (375), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 153/564 (27%), Positives = 240/564 (43%), Gaps = 81/564 (14%)
Query 4 VGTIKYLAKRCYHGKNVINPYTGQVMYQPCGECPACLTRKASIRSMRVSLQKSLSKYTYF 63
+ I Y +CYH ++V N YTG+V+ CG C ACL R+A S ++++ KY F
Sbjct 1 MSDIGYPLVKCYHPRHVQNKYTGEVIQVGCGVCKACLKRRADKMSFLCAIEEQSHKYCMF 60
Query 64 INPSYDQKYVPKCQIFKVHPD-DPDSTL---YELVVKPRRRGDykmvqkvyskkykkvik 119
+Y YVP+ ++P+ D + L Y + +G
Sbjct 61 ATLTYSNDYVPR-----MYPEVDNELRLVRWYSYCDRLNEKGKLM--------------- 100
Query 120 kEVDAPLSYTDDFSYYFNASEEYIRDFREQATLNVNGKYPHLQDYYGYISRKDGQLFVKR 179
T D+ Y+ A N++G Y Y S++D QLF+KR
Sbjct 101 ---------TVDYDYWHKCPSLDTYVLMLTAKCNLDG-------YLSYTSKRDAQLFLKR 144
Query 180 LRKHIFKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDSDRVAKNIGRIVNSSWKFGRC 239
+RK++ K++ EK+ Y+VSEYGP FR H+H+L F D + K + +++ +W+FGR
Sbjct 145 VRKNLSKYSD--EKIRYYIVSEYGPKTFRAHYHVLFFYDEVKTQKVMSKVIRQAWQFGRV 202
Query 240 DWSASRGDAESYVAGYVNSFSRLPYHLKQDDRVKPYSRFSNGFATSCFFDAKEAV-RDSI 298
D S SRG SYVA YVN LP L D KP+S S FA KE + + S+
Sbjct 203 DCSLSRGKCNSYVARYVNCNYCLPRFLG-DMSTKPFSCHSIRFALGIHQSQKEEIYKGSV 261
Query 299 SRPIEETPLSPFLNGVPSLINGKLLAIRPPRSVVDSCFLRYACNG--RLSSHEL---YWL 353
I ++ ING + P R++ SC C G R S EL Y +
Sbjct 262 DDFIYQS----------GEINGNYVEFMPWRNL--SCTFFPKCKGYSRKSDSELWQSYNI 309
Query 354 VRSVSTTLSRSFQTVRRDNPDATLMDVCRLHLRSIYSMSSRSVEDFLSIENQLYTCYYYA 413
+R V + + SF T+ + ++D L + + +S SR + N++ + Y
Sbjct 310 LREVRSAIGYSFNTII--DYARCILD---LVVTAKFSCDSRGLPCSSPALNKVIS---YF 361
Query 414 RLESPTTDVRSDDLFDSDCMRLYRLFSCVGKFISFWDITPSSSYEYIYRTVDLSKWYYSR 473
T SD L D + R F++F + + SY YR L + ++ R
Sbjct 362 SQGIDTNPYYSDYLADYHTNSIARELYISRHFLTF--VCDNDSYHERYRKFTLIRQFWQR 419
Query 474 LSYHMLRSQYTdlvglvdldeelvdFLIAPVTEDTPSSNGVSALEHHPPFESLVKSHPIL 533
Y +L YT + L + I D+ + VS L ++ V
Sbjct 420 YDYALLVGMYTSQIENRHLISNYDWYYINKTPLDSFGNVDVSQLSKELFYKRFV------ 473
Query 534 SLAQAKAKTDCDNRVKHREINERN 557
K+ + + +KH+ N+ N
Sbjct 474 ----IKSDENFEKSIKHKIQNDAN 493
>gi|496050828|ref|WP_008775335.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448892|gb|EEO54683.1| hypothetical protein BSCG_01608 [Bacteroides sp. 2_2_4]
Length=497
Score = 123 bits (308), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 84/274 (31%), Positives = 121/274 (44%), Gaps = 49/274 (18%)
Query 13 RCYHGKNVINPYTGQVMYQPCGECPACLTRKASIRSMRVSLQKSLSKYTYFINPSYDQKY 72
+C H K ++NPYT + M PCG C AC K S + + L+ +K+T FI +Y ++
Sbjct 8 KCLHPKRIMNPYTKESMVVPCGHCQACTLAKNSRYAFQCDLESYTAKHTLFITLTYANRF 67
Query 73 VPKCQIFKVHPDDPDSTLYELVVKPRRRGDykmvqkvyskkykkvikkEVDAPLSYTDDF 132
+P+ D Y + + G E+ P T+D
Sbjct 68 IPRAMFV-----DSIERPYGCDLIDKETG-------------------EILGPADLTED- 102
Query 133 SYYFNASEEYIRDFREQATLNVNGKYPHLQDYYGYISRKDGQLFVKRLRKHIFKFAGKYE 192
N+ K+ D Y+ + D QLF+KRLR ++ K E
Sbjct 103 -----------------ERTNLLNKFYLFGDV-PYLRKTDLQLFLKRLRYYVTK-QKPSE 143
Query 193 KVHIYLVSEYGPVHFRPHFHILLFTDSDRVAKNIGRIVNSSWKFGRCDWSASRGDAESYV 252
KV + V EYGPVHFRPH+H+LLF SD + ++ +W FGR D S+G +YV
Sbjct 144 KVRYFAVGEYGPVHFRPHYHLLLFLQSDEALQICSENISKAWTFGRVDCQVSKGQCSNYV 203
Query 253 AGYVNSFSRLPYHLKQDDRVKPYS----RFSNGF 282
A YVNS +P K V P+S + GF
Sbjct 204 ASYVNSSCTIPKVFKASS-VCPFSVHSQKLGQGF 236
>gi|494822887|ref|WP_007558295.1| hypothetical protein [Bacteroides plebeius]
gi|198272100|gb|EDY96369.1| hypothetical protein BACPLE_00805 [Bacteroides plebeius DSM 17135]
Length=545
Score = 110 bits (274), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 124/469 (26%), Positives = 189/469 (40%), Gaps = 108/469 (23%)
Query 14 CYHGKNVINPYTGQVMYQPCGECPACLTRKASIRSMRVSLQKSLSKYTYFINPSYDQKYV 73
C + + N YTG+ M C C AC + S + +K T F+ ++D K+V
Sbjct 8 CLEPQRIKNKYTGEEMVVACKHCVACEQLRNFKYSNLCDFESLTAKKTVFLTLTFDDKFV 67
Query 74 PKCQIFKVHPDDPDSTLYELVVKPRRRGDykmvqkvyskkykkvikkEVDAPLSYTDDFS 133
P+ + +KV D E +++ G+Y + P
Sbjct 68 PQFRFYKVGDD-------EYIMRDADTGEYLGRT--------------LMTP-------- 98
Query 134 YYFNASEEYIRDFREQATLNVNGKYPHLQDYYGYISRKDGQLFVKRLRKHIFKFAGKYEK 193
EY + R +N G++P Y+S+++ QLF+KRLRK++ K+ G +K
Sbjct 99 ---QLMNEYQK--RVNYRINYKGRFP-------YLSKRELQLFMKRLRKYLDKYEG--QK 144
Query 194 VHIYLVSEYGPVHFRPHFHILLFTD-----------------------------SDRVAK 224
+ + EYGP+ FRPHFHILLF D +
Sbjct 145 IRFFATGEYGPLSFRPHFHILLFVDDPSLFLPSVHTLGEYPYPYWSKYQKAHCGKGTLLS 204
Query 225 NIGRIVNSSWKFGRCD-WSASRGDAESYVAGYVNSFSRLPYHLKQDDRVKPYSRFSNGFA 283
+ + SW FG D S +G SYVAGYVNS LP LK D VK +S+ S
Sbjct 205 KLEYYIRESWPFGGIDAQSVEQGSCSSYVAGYVNSSVPLPSCLKV-DAVKSFSQHSRFLG 263
Query 284 TSCFFDAKEAVRDSISRPIEETPLSPFLNGVPSLINGKLLAIRPPRSVVDSCFLRYACNG 343
F E + P+ + + F+ G+ R P ++ S + + C G
Sbjct 264 RKIF--GTELI------PLLKLKFTEFVQR-SFFCRGRYDNFRTPSEMLHSVYPQ--CKG 312
Query 344 -RLSSHELYWLVRSVSTTLSRSFQTVRRDNPDATLMDVCRLHLRSIYS--------MSSR 394
L SHE + V ++ + L F + ++ DV R + S YS + R
Sbjct 313 FALLSHEQRFRVYTIWSRLRYYFNSDKK-------ADVARSLVTSFYSWLDTGILRVPER 365
Query 395 SVEDFLSIENQLYTCYYYARLESPTTD-VRSDDLFDSDCMRLYRLFSCV 442
EDFL I +L Y R++ D R DD ++ +LF CV
Sbjct 366 VREDFLLIYTELSQNLNYKRIDRFDYDKFRHDDDLNN------QLFQCV 408
>gi|575094322|emb|CDL65709.1| unnamed protein product [uncultured bacterium]
Length=499
Score = 105 bits (262), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 97/352 (28%), Positives = 148/352 (42%), Gaps = 63/352 (18%)
Query 32 PCGECPACLTRKASIRSMRVSLQKSLSKYTYFINPSYDQKYVPKCQIFKVHPDDPDSTLY 91
PCG+C AC K S S+++ L++ SKY YF+ +YD +P +F V D +
Sbjct 25 PCGKCIACHNNKRSSLSLKLRLEEYTSKYCYFLTLTYDDDNLP---LFSVGLDTCATEFV 81
Query 92 ELV-VKPRRRGDykmvqkvyskkykkvikkEVDAPLSYTDDF-SYYFNASEEYI--RDFR 147
+ R R D S+ DF S N +++ D+
Sbjct 82 RIYPYSERLRND------------------------SFISDFCSDLHNFDNDFVDKMDYY 117
Query 148 EQATLNVNGKYPHLQDYYGY-----ISRKDGQLFVKRLRKHIFKFAGKYEKVHIYLVSEY 202
+N KY H YG+ + +D QLF+KRLRKHI+K+ G EK+ Y++ EY
Sbjct 118 SDYVINYESKY-HKSCVYGHGLYALLYYRDIQLFLKRLRKHIYKYYG--EKIRFYIIGEY 174
Query 203 GPVHFRPHFHILLFTDSDRVAK------NIG---------RIVNSSWKFGRCDWSASRGD 247
G RPH+H LLF +S +++ N+G R + W+FG CD + G+
Sbjct 175 GTKSLRPHWHCLLFFNSSSLSQAFEDCVNVGTTSRPCSCPRFLRPFWQFGICDSKRTNGE 234
Query 248 AESYVAGYVNSFSRLPYHLKQDDRVKPYSRFSNGFATSCFFDAKEAVRDSISRPIEETPL 307
A +YV+ YVN + P L K Y G S SI I++
Sbjct 235 AYNYVSSYVNQSANFPKLLVLLSNQKAYHSIQLGQILS---------EQSIVSAIQKGDF 285
Query 308 SPFLNGVPSLINGKLLAIRPPRSVVDSCFLRYACNGRLSSHELYWLVRSVST 359
S F G + RS F ++ C+ +L+ + Y ++ T
Sbjct 286 SFFERQFYLDTFGAANSYSVWRSYYSRFFPKFTCSSQLTYEQTYRVLTCYET 337
>gi|575094355|emb|CDL65737.1| unnamed protein product [uncultured bacterium]
Length=517
Score = 104 bits (260), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 100/324 (31%), Positives = 137/324 (42%), Gaps = 75/324 (23%)
Query 13 RCYHGKNVINPYTGQVMYQPCGECPACLTRKASIRSMRVSLQKSLSKYTYFINPSYDQKY 72
RC K V NPY + PCG+C AC KAS +++ L+ S K+ F +Y Y
Sbjct 10 RCLEPKRVFNPYLNDWLLVPCGKCRACQCSKASRYKLQIQLEASQHKFCIFGTLTYANTY 69
Query 73 VPKCQIFKVHPDDPDSTLYELVVKPRRRGDykmvqkvyskkykkvikkEVDAPLSYTDDF 132
+P+ + + D T VV D E L Y D
Sbjct 70 IPRLSLVPYN----DKTF--GVVNGYEMCDK-----------------ETGEYLGYLDSP 106
Query 133 SYYFNASEEYIRDFREQATLNVNGKYPHLQDYYGYISRKDGQLFVKRLRKHIFKFAGKYE 192
SY E + D L++ G P Y+ ++D QLF+KRLRK++ K++
Sbjct 107 SY----DVESLLD-----KLHLFGDVP-------YLRKRDLQLFIKRLRKNLSKYSDA-- 148
Query 193 KVHIYLVSEYGPVHFRPHFHILLFTDSDRVAKNIGRI----------------------- 229
KV + + EYGPVHFRPH+H LLF D + G
Sbjct 149 KVRYFAMGEYGPVHFRPHYHFLLFFDEIKFTAPSGHTLGEFPDWAWYDSQNKCSRSDILS 208
Query 230 -----VNSSWKFGRCDWSASRGDAESYVAGYVNSFSRLPYHLKQDDRVKPY---SRF-SN 280
+ SSWKFGR D S+GDA YV+ YV+ LP + Q +P+ SRF
Sbjct 209 VVEYCIRSSWKFGRVDAQYSKGDAAQYVSSYVSGSGSLP-KVYQVSSARPFSLHSRFLGQ 267
Query 281 GF-ATSCFFDAKEAVRDSISRPIE 303
GF A C + VRD + R +E
Sbjct 268 GFLAHECEKVYETPVRDFVKRSVE 291
>gi|490418708|ref|WP_004291031.1| hypothetical protein [Bacteroides eggerthii]
gi|217986635|gb|EEC52969.1| hypothetical protein BACEGG_02720 [Bacteroides eggerthii DSM
20697]
Length=422
Score = 100 bits (250), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 51/104 (49%), Positives = 65/104 (63%), Gaps = 1/104 (1%)
Query 160 HLQDYYGYISRKDGQLFVKRLRKHIFKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDS 219
HL Y Y+ + D QLF KR R ++ K K EKV + + EYGPVHFRPH+HILLF S
Sbjct 36 HLFGYLPYLRKFDLQLFFKRFRYYVAKRFPK-EKVRYFAIGEYGPVHFRPHYHILLFLQS 94
Query 220 DRVAKNIGRIVNSSWKFGRCDWSASRGDAESYVAGYVNSFSRLP 263
D + ++V+ +W FGR D S+G SYVAGYVNS +P
Sbjct 95 DEALQVCSKVVSEAWPFGRVDCQLSKGKCSSYVAGYVNSSVLVP 138
>gi|490477382|ref|WP_004347759.1| hypothetical protein [Prevotella buccalis]
gi|281300711|gb|EFA93042.1| hypothetical protein HMPREF0650_1078 [Prevotella buccalis ATCC
35310]
Length=582
Score = 84.7 bits (208), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 88/367 (24%), Positives = 148/367 (40%), Gaps = 64/367 (17%)
Query 10 LAKRCYHGKNVINPYTGQVMYQPCGECPACLTRKASIRSMRVSLQKSLSKYTYFINPSYD 69
+ C + + V NP MY C +C ACL +KA+ S R + KY+ F +YD
Sbjct 9 IVGNCLNPRKVYNPSLHGWMYCSCDKCTACLNQKATTLSNRARAEIEQHKYSVFFTLTYD 68
Query 70 QKYVPKCQIFKVHPDDPDSTLYELVVKPRRRGDykmvqkvyskkykkvikkEVDAPLSYT 129
+++PK ++F+ DS V++ R G P++
Sbjct 69 NEHLPKYEVFQ------DSNE---VIQYRPIGRLVDDSSSDMLSN--------SCPINKY 111
Query 130 DDFSYYFNASEEYIRDFREQATLNVNGKYPHLQDY-----YGYISRKDGQLFVKRLRKHI 184
+++ E + F E + P +++Y +G + +KD Q F+KRLR I
Sbjct 112 NNY--------ENLYQFDESTFI------PPIENYEDIYHFGVVCKKDIQNFLKRLRWRI 157
Query 185 FKFAGKYE---KVHIYLVSEYGPVHFRPHFHILLFTDSDRVAKNIGRIVNSSW-KFGRCD 240
K + K+ Y+ SEYGP +RPH+H +LF DS ++ I ++ SW K+ R
Sbjct 158 SKIPNITKDESKIRYYISSEYGPTTYRPHYHGILFFDSKKILDKIKSLIVMSWGKYERQQ 217
Query 241 WSASR--------------------GDAESYVAGYVNSFSRLPYHLKQDDRVKPYSRFSN 280
+R + YVA YV LP L Q KP+ S
Sbjct 218 GERNRFKFRPFARPSLTSDYIKLCDPNTAYYVASYVAGNDNLPQVL-QLRETKPFHVQSK 276
Query 281 GFATSCFFDAKEAVRDSISRPIEETPLSPFLNGVPSLINGKLLAIRPPRSVVDSCFLRYA 340
+ K+ + ++I+R T F + + + + P + + S F +
Sbjct 277 NPVIGSYKVDKQEILENINRGTYTTTKQVFDDKTGQFND---VIVPLPETTLSSIFRKCV 333
Query 341 CNGRLSS 347
C L++
Sbjct 334 CYSTLTN 340
>gi|496521300|ref|WP_009229583.1| hypothetical protein [Prevotella sp. oral taxon 317]
gi|288330571|gb|EFC69155.1| hypothetical protein HMPREF0670_00478 [Prevotella sp. oral taxon
317 str. F0108]
Length=569
Score = 80.9 bits (198), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 60/227 (26%), Positives = 100/227 (44%), Gaps = 44/227 (19%)
Query 19 NVINPYTGQVMYQPCGECPACLTRKASIRSMRVSLQKSLSKYTYFINPSYDQKYVPKCQI 78
+V N +T M+ PCG C AC+ AS +S RV + KY+ +Y+ +++P+ +
Sbjct 16 HVHNRWTRDEMFVPCGRCEACVNAAASKQSKRVRNEIMQHKYSVMFTLTYNNEFIPRWER 75
Query 79 FKVHPDDPDSTLYELVVKPRRRGDykmvqkvyskkykkvikkEVDAPLSYTDDFSYYFNA 138
F + D P ++P R PL+Y D +
Sbjct 76 FLDNNDCPQ-------LRPIGRCAELFP----------------SCPLNYFDKVT----- 107
Query 139 SEEYIRDFREQATLNVNGKYPHLQ-----DYYGYISRKDGQLFVKRLRKHIFKFAGKYE- 192
+ +++++ P ++ + + +KD Q F+KRLR +I K GK E
Sbjct 108 ---------GKWSIDLDTFLPKIENDEHTEVFASCCKKDIQNFLKRLRFNISKLYGKAES 158
Query 193 -KVHIYLVSEYGPVHFRPHFHILLFTDSDRVAKNIGRIVNSSWKFGR 238
K+ Y+ SEYGP RPH+H ++F D + I ++ SW F R
Sbjct 159 RKIRYYVASEYGPTTLRPHYHGIIFFDDASLLSEISSLIVRSWGFQR 205
>gi|494610270|ref|WP_007368516.1| hypothetical protein [Prevotella multiformis]
gi|324988542|gb|EGC20505.1| hypothetical protein HMPREF9141_0984 [Prevotella multiformis
DSM 16608]
Length=479
Score = 80.5 bits (197), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 56/176 (32%), Positives = 84/176 (48%), Gaps = 22/176 (13%)
Query 165 YGYISRKDGQLFVKRLRK----HIFKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDSD 220
+ Y +KD Q + KRLR + K ++ ++ SEYGP FRPH+H +L+ DS+
Sbjct 116 FAYPCKKDVQDWFKRLRSAVDYQLNKNKSNEFRIRYFICSEYGPRTFRPHYHAILWYDSE 175
Query 221 RVAKNIGRIVNSSWKFGRCDWSASRGDAESYVAGYVNSFSRLPYHLKQDDRVKPYSRFSN 280
+ +NIGR++ +WK G +S A YVA YVN +RLP L+ +
Sbjct 176 ELQRNIGRLIRETWKNGNSVFSLVNNSASQYVAKYVNGDTRLPPFLRTE----------- 224
Query 281 GFATSCFFDAKEAVRDSISRPIEETPLSPFLNG-----VPSLINGKLLAIRPPRSV 331
TS F A + + EE S L+G V + NG+ + PRS+
Sbjct 225 --FTSTFHLASKHPYIGYCKADEEALRSNVLDGTYGQSVLNRDNGQFEFVPTPRSL 278
Score = 39.3 bits (90), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 36/77 (47%), Gaps = 4/77 (5%)
Query 1 MDKVGTIKYLAKRCYHGKNVINPYTGQVMYQPCGECPACLTRKASIRSMRVSLQKSLSKY 60
M +G I C K + N Y + +Y PC +C C AS S R+ + ++
Sbjct 1 MSVLGNIN----SCLSPKRIYNKYIDETLYVPCRKCFRCRDSYASDWSRRIENECREHRF 56
Query 61 TYFINPSYDQKYVPKCQ 77
+ F+ +YD +++P Q
Sbjct 57 SLFVTLTYDNEHIPLFQ 73
>gi|647452984|ref|WP_025792805.1| hypothetical protein [Prevotella histicola]
Length=480
Score = 75.1 bits (183), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 40/108 (37%), Positives = 58/108 (54%), Gaps = 3/108 (3%)
Query 165 YGYISRKDGQLFVKRLRKHI---FKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDSDR 221
+ Y +KD Q F KRLR I K G ++ ++ SEYGP FRPH+H +L+ DS+
Sbjct 119 FAYPCKKDVQDFFKRLRSKIDYKLKPRGNEYRIRYFICSEYGPNTFRPHYHAILWYDSEI 178
Query 222 VAKNIGRIVNSSWKFGRCDWSASRGDAESYVAGYVNSFSRLPYHLKQD 269
+ + ++ +WK G D+S A YVA YVN LP L+ +
Sbjct 179 LHNELNVLIRETWKNGNTDFSLVNSSASQYVAKYVNGDCDLPSFLRTE 226
Lambda K H a alpha
0.322 0.137 0.422 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4106350928880