bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-39_CDS_annotation_glimmer3.pl_2_1
Length=322
Score E
Sequences producing significant alignments: (Bits) Value
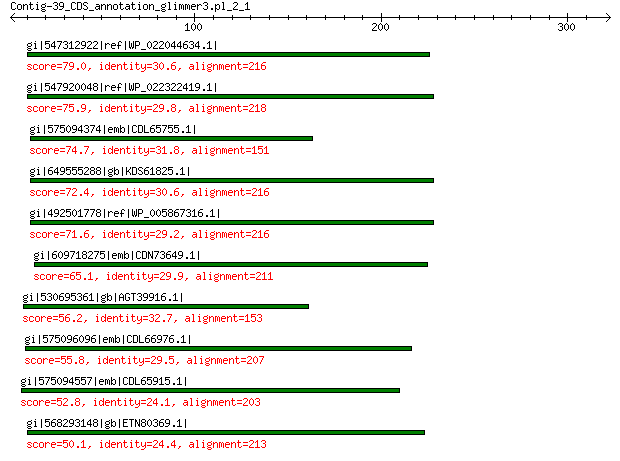
gi|547312922|ref|WP_022044634.1| putative replication initiation... 79.0 2e-13
gi|547920048|ref|WP_022322419.1| putative replication protein 75.9 1e-12
gi|575094374|emb|CDL65755.1| unnamed protein product 74.7 1e-11
gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 72.4 2e-11
gi|492501778|ref|WP_005867316.1| hypothetical protein 71.6 5e-11
gi|609718275|emb|CDN73649.1| conserved hypothetical protein 65.1 6e-09
gi|530695361|gb|AGT39916.1| replication initiator 56.2 9e-06
gi|575096096|emb|CDL66976.1| unnamed protein product 55.8 1e-05
gi|575094557|emb|CDL65915.1| unnamed protein product 52.8 1e-04
gi|568293148|gb|ETN80369.1| hypothetical protein NECAME_18023 50.1 0.001
>gi|547312922|ref|WP_022044634.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
gi|524208442|emb|CCZ76638.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
Length=320
Score = 79.0 bits (193), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 66/229 (29%), Positives = 105/229 (46%), Gaps = 33/229 (14%)
Query 10 VEVECGHCFECRKKKRREWRIRNYEQLKETP--IAVFFTGTVSPQRYEHICKQYGYKNDG 67
+EV CG+C C+K ++RIR +L++ P +F T T + E K
Sbjct 40 LEVPCGYCHSCQKSYNNQYRIRLLYELRKYPPGTCLFVTLTFNDDSLEKFSKD------- 92
Query 68 SQDNEIITKIQRLFLERIRKEKGYSIKHWCVTEKGHTNTRRIHIHGLYYAT-------HG 120
K RLFL+R RK G I+HW V E G T R H HG+ + +
Sbjct 93 ------TNKAVRLFLDRFRKVYGKQIRHWFVCEFG-TLHGRPHYHGILFNVPQALIDGYD 145
Query 121 ETKWQLTKTLFENWIDGYRFYGSYVNEKTINYVSKYMTKK---DEDNPDYIGIVLCSKGL 177
L W G+ F G YV+++T +Y++KY+TK D+ P V+ S G+
Sbjct 146 SDMPGHHPLLASCWKYGFVFVG-YVSDETCSYITKYVTKSINGDKVRPR----VISSFGI 200
Query 178 GANYAK-RMAYKHEWNKEKTNITYKAKNGADLPLPRYYKTQLYTEDQRQ 225
G+NY + H+ ++ + NG +PRYY +++++ +Q
Sbjct 201 GSNYLNTEESSLHKLGNQRYQ-PFMVLNGFQQAMPRYYYNKIFSDVDKQ 248
>gi|547920048|ref|WP_022322419.1| putative replication protein [Parabacteroides merdae CAG:48]
gi|524592960|emb|CDD13572.1| putative replication protein [Parabacteroides merdae CAG:48]
Length=278
Score = 75.9 bits (185), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 65/228 (29%), Positives = 106/228 (46%), Gaps = 18/228 (8%)
Query 10 VEVECGHCFECRKKKRREWRIRNYEQLKETPIAVFFTGTVSPQRYEHI-CKQYGYKNDGS 68
+V CG C CR+ KR+ W R + KE P+++F T T EH+ ++ G +D
Sbjct 8 AKVPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDD---EHLPIERIG--SDLF 62
Query 69 QDNEIITKIQ--RLFLERIRKE-KGYSIKHWCVTEKGHTNTRRIHIHGLYYATHGETKWQ 125
Q N + + +LF++R+RK+ + Y ++++ +E G N R H H + + K
Sbjct 63 QTNVAVVSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRP-HYHMILFGFPFTGK-M 120
Query 126 LTKTLFENWIDGYRFYGSYVNEKTINYVSKYMTKKD------EDNPDYIGIVLCSKGLGA 179
L E W +G+ + K I YV KYM +K D Y +LCS+ G
Sbjct 121 AGDLLAECWQNGF-VQAHPLTIKEIAYVCKYMYEKSMCPEILRDEKKYKPFMLCSRNPGI 179
Query 180 NYAKRMAYKHEWNKEKTNITYKAKNGADLPLPRYYKTQLYTEDQRQLL 227
+ A E+ + +A G + +PRYY +LY +D + L
Sbjct 180 GFGFMKADIIEFYRRHPRDYVRAWAGHKMAMPRYYADKLYDDDMKAFL 227
>gi|575094374|emb|CDL65755.1| unnamed protein product [uncultured bacterium]
Length=487
Score = 74.7 bits (182), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 48/158 (30%), Positives = 76/158 (48%), Gaps = 12/158 (8%)
Query 12 VECGHCFECRKKKRREWRIRNYEQLKETPIAVFFTGTVSPQRYEHICKQYGYKNDGSQDN 71
V CGHC++C+ K +W++R E+L + F+T T+ P+ YG DGS
Sbjct 26 VPCGHCYDCKSAKTTDWQVRCSEELNNNSQSYFYTLTLDPR----FIDTYGTLPDGSPRY 81
Query 72 EIITKIQRLFLERIRKEKG---YSIKHWCVTEKGHTNTRRIHIHGLYYATHGETKWQLTK 128
+ +LFL+R+RK S+K+ V E G T T R H H ++Y + ++
Sbjct 82 VFNKRHIQLFLKRLRKALSKYNISLKYVIVGELGET-THRPHYHAIFYLSSSVNPFKFRI 140
Query 129 TLFENWIDGYRFYGS----YVNEKTINYVSKYMTKKDE 162
+ +W G+ G +N ++YV KYM K D
Sbjct 141 MVRNSWSLGFIKSGDNNGIILNNDAVSYVIKYMHKTDS 178
>gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649560564|gb|KDS66872.1| hypothetical protein M095_2449 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561011|gb|KDS67298.1| hypothetical protein M095_2409 [Parabacteroides distasonis str.
3999B T(B) 4]
Length=284
Score = 72.4 bits (176), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 66/226 (29%), Positives = 104/226 (46%), Gaps = 18/226 (8%)
Query 12 VECGHCFECRKKKRREWRIRNYEQLKETPIAVFFTGTVSPQRYEHICKQYGYKNDGSQDN 71
V CG C CRK KR+ W R + E P ++F T T EHI ++
Sbjct 15 VPCGRCVNCRKNKRQSWVYRLQAEADEYPFSLFVTLTYDD---EHIPTAMIGEDLFKTTV 71
Query 72 EIITK--IQRLFLERIRKEKG-YSIKHWCVTEKGHTNTRRIHIHGLYYATHGETKWQLTK 128
+++K IQ LF++R+RK+ Y ++++ +E G + R H H + + K
Sbjct 72 GVVSKRDIQ-LFMKRLRKKYAQYRLRYFLTSEYG-SQGGRPHYHMILFGFPFTGK-HGGD 128
Query 129 TLFENWIDGYRFYGSYVNEKTINYVSKYMTKKDEDNPD-------YIGIVLCSKGLGANY 181
L E W +G+ + K I+YV+KYM +K PD Y +LCSK G Y
Sbjct 129 LLAECWKNGF-VQAHPLTTKEISYVTKYMYEKSM-IPDILKGVKEYQPFMLCSKMPGIGY 186
Query 182 AKRMAYKHEWNKEKTNITYKAKNGADLPLPRYYKTQLYTEDQRQLL 227
++ + +A NG + +PRYY +LY +D ++ L
Sbjct 187 HFLREQILDFYRLHPRDYVRAFNGMRMAMPRYYADKLYDDDMKEYL 232
>gi|492501778|ref|WP_005867316.1| hypothetical protein [Parabacteroides distasonis]
gi|409230407|gb|EKN23271.1| hypothetical protein HMPREF1059_03256 [Parabacteroides distasonis
CL09T03C24]
Length=284
Score = 71.6 bits (174), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 63/225 (28%), Positives = 104/225 (46%), Gaps = 16/225 (7%)
Query 12 VECGHCFECRKKKRREWRIRNYEQLKETPIAVFFTGTVSPQRYEHICKQYGYKNDGSQDN 71
V CG C CRK KR+ W R + E P ++F T T EH+ ++
Sbjct 15 VPCGRCVNCRKNKRQSWVYRLQAEADEYPFSLFVTLTYDD---EHMPTAMIGEDLFKSTV 71
Query 72 EIITK--IQRLFLERIRKE-KGYSIKHWCVTEKGHTNTRRIHIHGLYYATHGETKWQLTK 128
+++K IQ LF++R+RK+ Y ++++ +E G + R H H + + K
Sbjct 72 GVVSKRDIQ-LFMKRLRKKYDQYRLRYFLTSEYG-SQGGRPHYHMILFGFPFTGK-HGGD 128
Query 129 TLFENWIDGYRFYGSYVNEKTINYVSKYMTKKD------EDNPDYIGIVLCSKGLGANYA 182
L E W +G+ + K I YV+KYM +K +D +Y +LCS+ G Y
Sbjct 129 LLAECWKNGF-VQAHPLTTKEIAYVTKYMYEKSMVPDILKDVKEYQPFMLCSRIPGIGYH 187
Query 183 KRMAYKHEWNKEKTNITYKAKNGADLPLPRYYKTQLYTEDQRQLL 227
++ + +A NG + +PRYY +LY +D ++ L
Sbjct 188 FLREQILDFYRLHPRDYVRAFNGMRMAMPRYYADKLYDDDMKEYL 232
>gi|609718275|emb|CDN73649.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=265
Score = 65.1 bits (157), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 63/221 (29%), Positives = 95/221 (43%), Gaps = 32/221 (14%)
Query 14 CGHCFECRKKKRREWRIRNYEQLKETPIAVFFTGTVSPQRYEHICKQYGYKNDGSQDNEI 73
CG C ECRK + W R E+LK + A F T T Y + Y DN +
Sbjct 25 CGKCLECRKARTNSWFARLTEELKVSKSAHFVTLT-----YSDVYLPY-------SDNGL 72
Query 74 ITKIQR---LFLERIRKEKGYSIKHWCVTEKGHTNTRRIHIHGLYYATHGETKWQLTKTL 130
I+ R LF++R RK + IK++ V E G T R H H + +
Sbjct 73 ISLDYRDFQLFMKRARKLQKSKIKYFLVGEYG-AQTYRPHYHAIVFGVEN------IDAF 125
Query 131 FENWIDGYRFYGSYVNEKTINYVSKYMTKKDEDNPDYIGI-------VLCSKGLGANYAK 183
W G G+ V K+I Y KY TK + PD L SKGLG ++
Sbjct 126 LGEWRMGNVHAGT-VTAKSIYYTLKYCTKSITEGPDKDPDDDRKPEKALMSKGLGLSHLT 184
Query 184 RMAYKHEWNKEKTNITYKAKNGADLPLPRYYKTQLYTEDQR 224
K + K+ + ++ G + LPRYY+ +++++ ++
Sbjct 185 ESMIK--YYKDDVSRSFSLLGGTTIALPRYYRDKVFSDIEK 223
>gi|530695361|gb|AGT39916.1| replication initiator [Marine gokushovirus]
Length=289
Score = 56.2 bits (134), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 50/163 (31%), Positives = 76/163 (47%), Gaps = 22/163 (13%)
Query 8 RYVEVECGHCFECRKKKRREWRIRNYEQLKETPIAVFFTGTVSPQRYEHICKQYGYKNDG 67
R + CG C CR R+W IR + + F T T EHI K+ KN
Sbjct 26 RGFNLPCGQCIGCRLDYSRQWAIRCVHEAQTHEDNCFITLTFDN---EHIAKR---KNPE 79
Query 68 SQDNEIITKIQRLFLERIRKEKGYSIKHWCVTEKGHTNTRRIHIHGLYY----------A 117
S DN T+ QR F++R+RK+ + I+ + E G N +R H H L + +
Sbjct 80 SLDN---TEFQR-FMKRLRKKYPHKIRFFHCGEYGDQN-KRPHYHALLFGHDFKDKKLWS 134
Query 118 THGETKWQLTKTLFENWIDGYRFYGSYVNEKTINYVSKYMTKK 160
G+ K +++ L E W G+ G+ V+ T Y ++Y+ KK
Sbjct 135 NKGDFKLFVSQELAELWPYGFHTIGA-VSFDTAAYCARYVMKK 176
>gi|575096096|emb|CDL66976.1| unnamed protein product [uncultured bacterium]
Length=296
Score = 55.8 bits (133), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 61/236 (26%), Positives = 101/236 (43%), Gaps = 52/236 (22%)
Query 9 YVEVECGHCFECRKKKRREWRIRNYEQLKETPIAVFFTGTVSPQRYEHICKQYGYKNDGS 68
YV V CG C ECR + EW +R +LK +F T T Y +D
Sbjct 33 YVLVPCGQCLECRLHRASEWALRCCHELKSHDKGIFLTLT--------------YNDDNL 78
Query 69 QDN-EIITKIQRLFLERIRKEKGY-----SIKHWCVTEKGHTNTRRIHIHGLYYATH--- 119
N ++ K + F++R+R+ Y I++ C E G + R H H L + +
Sbjct 79 PPNGTLVKKHVQDFIKRLRRHIDYYGDCTKIRYLCAGEYGDLSLRP-HYHLLVFGYYPSD 137
Query 120 ----------GETKWQLTKTLFENWIDGYRFYGSYVNEKTINYVSKYMTKKD--EDNPDY 167
G+ + TL + W G+ +G+ + ++ Y +Y KK E + Y
Sbjct 138 PRLLHGLQKIGKNSLFTSPTLTKLWGKGHISFGA-ITFESARYTCQYALKKQTGEHSHYY 196
Query 168 I--GIV----LCS--KGLGANYAKRMAYKHEWNKEKTNITYKAKNGADLPLPRYYK 215
+ G++ +CS GLG ++ A H+ E+ +T NG + +PRYY+
Sbjct 197 VDRGVIPEFMICSNRNGLGYDF----AVSHDNMFERGYLT---MNGKKIGIPRYYQ 245
>gi|575094557|emb|CDL65915.1| unnamed protein product [uncultured bacterium]
Length=354
Score = 52.8 bits (125), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 49/218 (22%), Positives = 93/218 (43%), Gaps = 19/218 (9%)
Query 7 FRYVEVECGHCFECRKKKRREWRIRNYEQLKETPIAVFFTGTVSPQRYEHICKQYGYKND 66
R V CG C CR K RREW R ++ F T T S + Y + + G++
Sbjct 25 LRGVPFGCGKCLACRVKTRREWTSRLILEMLGHDSGAFVTLTYS-EDYVPVTES-GHRTL 82
Query 67 GSQDNEIITKIQRLFLERIRKEKGYSIKHWCVTEKGHTNTRRIHIHGLYYATHGETKWQL 126
+D ++ K R LE RK + I+++ E G T+R H H +++ +
Sbjct 83 SLRDLQLFLKRLRRNLEE-RKRSKHPIRYYACGEYGTRGTQRPHYHIIFFGV-SDLDLDF 140
Query 127 TKTLFENWIDGYRF--------YGSY----VNEKTINYVSKYMTKKDEDNPDYIGIVLCS 174
K+++ W + ++ +G+ +N KT+ Y + Y KK +V+ S
Sbjct 141 IKSVYAAWSEPAKYGQKGQTPQFGNITIEPLNAKTVAYTAGYNMKKLISPKKVHKVVVSS 200
Query 175 KGLGA---NYAKRMAYKHEWNKEKTNITYKAKNGADLP 209
+G+ + K + + N++ + + + + +P
Sbjct 201 AEIGSRRVTFEKLVLDRKNSNRDDNGVLAEFRVMSRMP 238
>gi|568293148|gb|ETN80369.1| hypothetical protein NECAME_18023 [Necator americanus]
Length=345
Score = 50.1 bits (118), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 52/222 (23%), Positives = 95/222 (43%), Gaps = 25/222 (11%)
Query 10 VEVECGHCFECRKKKRREWRIRNYEQLKETPIAVFFTGTVSPQRYEHICKQYGYKNDGSQ 69
V V CG C C++++ W R ++ + A F T T R+ I K D +
Sbjct 18 VPVPCGRCPPCKRRRVDSWVFRLLQEELQHENASFVTLTYD-TRFVPISKNGFMTLDRGE 76
Query 70 DNEIITKIQRLFLERIRKEKGYSIKHWCVTEKGHTNTRRIHIHGLYYATHGETKWQLTKT 129
+ ++++L G +K++ E G + R H H + + ++ + T
Sbjct 77 FPRYMKRLRKLV-------PGRKLKYYMCGEYG-SQRFRPHYHAIIFGVPQDSLFADAWT 128
Query 130 LFENWIDGYRFYGSYVNEKTINYVSKYMTK--------KDEDNPDYIGIVLCSKGLGANY 181
L N V K+I Y KY+ K +D+ P++ L SKG+G +Y
Sbjct 129 L--NGDSLGGVVVGTVTGKSIAYTMKYIDKSTWKQKHGRDDRVPEF---SLMSKGMGVSY 183
Query 182 AKRMAYKHEWNKEKTNITYKAKNGAD-LPLPRYYKTQLYTED 222
E++KE + + + G + +PRYY+ ++Y++D
Sbjct 184 LTPQMV--EYHKEDISRLFCTREGGSRIAMPRYYRQKIYSDD 223
Lambda K H a alpha
0.318 0.136 0.422 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1793877651450