bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_5
Length=217
Score E
Sequences producing significant alignments: (Bits) Value
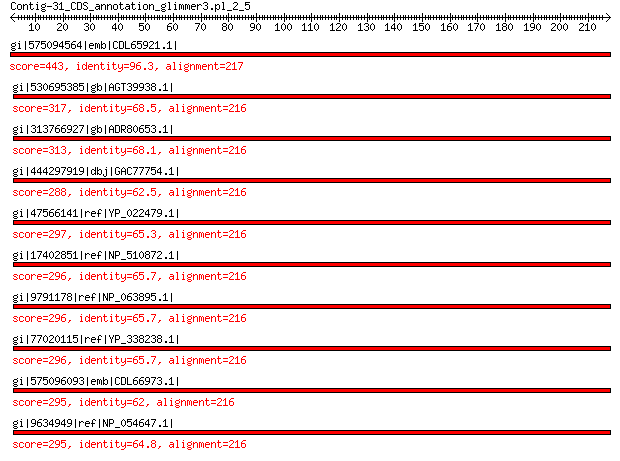
gi|575094564|emb|CDL65921.1| unnamed protein product 443 1e-150
gi|530695385|gb|AGT39938.1| major capsid protein 317 2e-102
gi|313766927|gb|ADR80653.1| putative major coat protein 313 8e-101
gi|444297919|dbj|GAC77754.1| major capsid protein 288 3e-94
gi|47566141|ref|YP_022479.1| structural protein 297 5e-94
gi|17402851|ref|NP_510872.1| hypothetical protein PhiCPG1p2 296 5e-94
gi|9791178|ref|NP_063895.1| hypothetical protein 296 6e-94
gi|77020115|ref|YP_338238.1| putative major coat protein 296 1e-93
gi|575096093|emb|CDL66973.1| unnamed protein product 295 3e-93
gi|9634949|ref|NP_054647.1| structural protein 295 3e-93
>gi|575094564|emb|CDL65921.1| unnamed protein product [uncultured bacterium]
Length=582
Score = 443 bits (1139), Expect = 1e-150, Method: Compositional matrix adjust.
Identities = 209/217 (96%), Positives = 214/217 (99%), Gaps = 0/217 (0%)
Query 1 MFNVISPDARLQRPEYLGGTHSRVNVVPTAQTSSTDSVSPQSNLSAFGVLGDSAHGFNKS 60
MFNVISPDARLQRPEYLGGTHSRVNVVPTAQTSSTDSVSPQSNLSAFGVLGDSAHGFNKS
Sbjct 366 MFNVISPDARLQRPEYLGGTHSRVNVVPTAQTSSTDSVSPQSNLSAFGVLGDSAHGFNKS 425
Query 61 FVEHGYVIGLCCLRADITYQQGLNRMWSRRQLFDFYWPTLAHLGEQVVYNKEIYAQGTAD 120
FVEHGYVIGL CLRADITYQQGLNRMWSRRQLFDFYWPTLAHLGEQVVYN+EIY QGT D
Sbjct 426 FVEHGYVIGLVCLRADITYQQGLNRMWSRRQLFDFYWPTLAHLGEQVVYNREIYTQGTDD 485
Query 121 DNGVFGYQERYAEYRYKPSMITGKLRSTDAQSLDVWHLAQRFDSLPKLNQDFIEENPPIN 180
DNGVFGYQERYAEYRYKPSMITGKLRSTD+Q+LDVWHLAQ+FD+LPKLNQDFIEENPPIN
Sbjct 486 DNGVFGYQERYAEYRYKPSMITGKLRSTDSQTLDVWHLAQKFDTLPKLNQDFIEENPPIN 545
Query 181 RVIAVQNEPQFFADFWFDLKTSRPMPVYSVPGLVDHF 217
RVIAVQNEPQFFADFWFDLKTSRPMPVYSVPGLVDHF
Sbjct 546 RVIAVQNEPQFFADFWFDLKTSRPMPVYSVPGLVDHF 582
>gi|530695385|gb|AGT39938.1| major capsid protein [Marine gokushovirus]
Length=514
Score = 317 bits (813), Expect = 2e-102, Method: Compositional matrix adjust.
Identities = 148/216 (69%), Positives = 168/216 (78%), Gaps = 0/216 (0%)
Query 2 FNVISPDARLQRPEYLGGTHSRVNVVPTAQTSSTDSVSPQSNLSAFGVLGDSAHGFNKSF 61
F V SPDARLQRPEYLGG R+N+ P AQTSSTD+ +PQ NLS +G G + H FNKSF
Sbjct 299 FGVTSPDARLQRPEYLGGGKDRININPIAQTSSTDATTPQGNLSGYGTTGFTGHRFNKSF 358
Query 62 VEHGYVIGLCCLRADITYQQGLNRMWSRRQLFDFYWPTLAHLGEQVVYNKEIYAQGTADD 121
EH V+GL C+ AD+TYQQGL R +SR+ +DFYWP LAHLGEQ V NKEIYAQGT DD
Sbjct 359 TEHSVVLGLACVFADLTYQQGLPRHFSRQTRWDFYWPALAHLGEQAVLNKEIYAQGTTDD 418
Query 122 NGVFGYQERYAEYRYKPSMITGKLRSTDAQSLDVWHLAQRFDSLPKLNQDFIEENPPINR 181
N VFGYQERYAEYRYKPS ITG++RS AQSLD+WHLAQ F SLP LN FIEENPP++R
Sbjct 419 NNVFGYQERYAEYRYKPSSITGQMRSNFAQSLDIWHLAQDFGSLPVLNSSFIEENPPVDR 478
Query 182 VIAVQNEPQFFADFWFDLKTSRPMPVYSVPGLVDHF 217
V AVQN P D +F LK +RPMP Y VPGL+DHF
Sbjct 479 VTAVQNYPNLILDMYFKLKCARPMPTYGVPGLIDHF 514
>gi|313766927|gb|ADR80653.1| putative major coat protein [Uncultured Microviridae]
Length=533
Score = 313 bits (803), Expect = 8e-101, Method: Compositional matrix adjust.
Identities = 147/216 (68%), Positives = 171/216 (79%), Gaps = 1/216 (0%)
Query 2 FNVISPDARLQRPEYLGGTHSRVNVVPTAQTSSTDSVSPQSNLSAFGVLGDSAHGFNKSF 61
F V SPDARLQRPEYLGG + V + QTSSTDS SPQ NL+A G S GF+KSF
Sbjct 304 FGVTSPDARLQRPEYLGGQKTEVMMQTVPQTSSTDSTSPQGNLAALGT-ATSRGGFSKSF 362
Query 62 VEHGYVIGLCCLRADITYQQGLNRMWSRRQLFDFYWPTLAHLGEQVVYNKEIYAQGTADD 121
VEHG +IGL C+ AD+TYQQG+NRMWSRR +DFYWP+LAHLGEQ V N+EIY QGT+ D
Sbjct 363 VEHGVLIGLACVFADLTYQQGMNRMWSRRDRWDFYWPSLAHLGEQAVLNQEIYTQGTSAD 422
Query 122 NGVFGYQERYAEYRYKPSMITGKLRSTDAQSLDVWHLAQRFDSLPKLNQDFIEENPPINR 181
FGYQER+AEYRYKPS ITGK+RS +LD WHLAQ F +LP LN FIEENPP++R
Sbjct 423 TQTFGYQERFAEYRYKPSQITGKMRSNATGTLDAWHLAQDFTALPALNASFIEENPPVDR 482
Query 182 VIAVQNEPQFFADFWFDLKTSRPMPVYSVPGLVDHF 217
VIAV +EP+F D++FDLKT+RPMPVYSVPGL+DHF
Sbjct 483 VIAVPSEPEFIWDWYFDLKTTRPMPVYSVPGLIDHF 518
>gi|444297919|dbj|GAC77754.1| major capsid protein [uncultured marine virus]
Length=283
Score = 288 bits (738), Expect = 3e-94, Method: Compositional matrix adjust.
Identities = 135/217 (62%), Positives = 167/217 (77%), Gaps = 1/217 (0%)
Query 2 FNVISPDARLQRPEYLGGTHSRVNVVPTAQTSSTDSV-SPQSNLSAFGVLGDSAHGFNKS 60
F V SPD+RLQRPEYLGG S V ++P AQTS +++ + Q L+A G S GF KS
Sbjct 67 FKVESPDSRLQRPEYLGGGSSLVQILPIAQTSQSEATGTEQGKLTAVGYHSQSGLGFTKS 126
Query 61 FVEHGYVIGLCCLRADITYQQGLNRMWSRRQLFDFYWPTLAHLGEQVVYNKEIYAQGTAD 120
FVEH +IGL +RAD+TYQQG++RMWSR+ +DFYWP LA+LGEQ V NKEI+ Q A
Sbjct 127 FVEHCVIIGLVNVRADLTYQQGMDRMWSRKTKYDFYWPALANLGEQTVLNKEIFTQAIAA 186
Query 121 DNGVFGYQERYAEYRYKPSMITGKLRSTDAQSLDVWHLAQRFDSLPKLNQDFIEENPPIN 180
D+ VFGYQER+AEYRY PS ITG LRS A SLD+WHL+Q F SLP LN+ FI+ENPP++
Sbjct 187 DDEVFGYQERWAEYRYFPSRITGVLRSDAAASLDLWHLSQDFGSLPALNESFIQENPPVD 246
Query 181 RVIAVQNEPQFFADFWFDLKTSRPMPVYSVPGLVDHF 217
RV+AV +EP+F D +FDL T+RPMP+YSVPGL+DHF
Sbjct 247 RVVAVTDEPEFIFDSYFDLITTRPMPMYSVPGLIDHF 283
>gi|47566141|ref|YP_022479.1| structural protein [Chlamydia phage 3]
gi|47522476|emb|CAD79477.1| structural protein [Chlamydia phage 3]
Length=565
Score = 297 bits (760), Expect = 5e-94, Method: Compositional matrix adjust.
Identities = 141/224 (63%), Positives = 165/224 (74%), Gaps = 8/224 (4%)
Query 2 FNVISPDARLQRPEYLGGTHSRVNVVPTAQTSSTDSVSPQSNLSAFGVLGDSAHGFNKSF 61
FNV SPDARLQR EYLGG+ + VN+ P QTSSTDS SPQ NL+A+G S F KSF
Sbjct 342 FNVQSPDARLQRAEYLGGSSTPVNISPIPQTSSTDSTSPQGNLAAYGTAIGSKRVFTKSF 401
Query 62 VEHGYVIGLCCLRADITYQQGLNRMWSRRQLFDFYWPTLAHLGEQVVYNKEIYAQGTADD 121
EHG ++GL +RAD+ YQQGL+RMWSRR +DFYWP L+HLGEQ V NKEIY QG +
Sbjct 402 TEHGVILGLASVRADLNYQQGLDRMWSRRTRWDFYWPALSHLGEQAVLNKEIYCQGPSVK 461
Query 122 NG--------VFGYQERYAEYRYKPSMITGKLRSTDAQSLDVWHLAQRFDSLPKLNQDFI 173
N VFGYQER+AEYRYK S ITGK RS SLD WHLAQ F++LP L+ +FI
Sbjct 462 NSGGEIVDEQVFGYQERFAEYRYKTSKITGKFRSNATSSLDSWHLAQEFENLPTLSPEFI 521
Query 174 EENPPINRVIAVQNEPQFFADFWFDLKTSRPMPVYSVPGLVDHF 217
EENPP++RV+AV NEP F D WF L+ +RPMPVYSVPG +DHF
Sbjct 522 EENPPMDRVLAVSNEPHFLLDGWFSLRCARPMPVYSVPGFIDHF 565
>gi|17402851|ref|NP_510872.1| hypothetical protein PhiCPG1p2 [Guinea pig Chlamydia phage]
Length=553
Score = 296 bits (759), Expect = 5e-94, Method: Compositional matrix adjust.
Identities = 142/226 (63%), Positives = 166/226 (73%), Gaps = 10/226 (4%)
Query 2 FNVISPDARLQRPEYLGGTHSRVNVVPTAQTSSTDSVSPQSNLSAFGVLGDSAHGFNKSF 61
FNV SPDARLQR EYLGG+ + VN+ P QTSSTDS SPQ NL+A+G S F KSF
Sbjct 328 FNVQSPDARLQRAEYLGGSSTPVNISPIPQTSSTDSTSPQGNLAAYGTAIGSKRVFTKSF 387
Query 62 VEHGYVIGLCCLRADITYQQGLNRMWSRRQLFDFYWPTLAHLGEQVVYNKEIYAQGTA-- 119
EHG ++GL +RAD+ YQQGL+RMWSRR +DFYWP L+HLGEQ V NKEIY QG A
Sbjct 388 TEHGVILGLASVRADLNYQQGLDRMWSRRTRWDFYWPALSHLGEQAVLNKEIYCQGPAVK 447
Query 120 --------DDNGVFGYQERYAEYRYKPSMITGKLRSTDAQSLDVWHLAQRFDSLPKLNQD 171
D VFGYQER+AEYRYK S ITGK RS SLD WHLAQ+F++LP L+ +
Sbjct 448 DAQNGNVVVDEQVFGYQERFAEYRYKTSKITGKFRSNATGSLDAWHLAQQFENLPTLSPE 507
Query 172 FIEENPPINRVIAVQNEPQFFADFWFDLKTSRPMPVYSVPGLVDHF 217
FIEENPP++RV+AV EP F D WF L+ +RPMPVYSVPGL+DHF
Sbjct 508 FIEENPPMDRVVAVDTEPDFLLDGWFSLRCARPMPVYSVPGLIDHF 553
>gi|9791178|ref|NP_063895.1| hypothetical protein [Chlamydia pneumoniae phage CPAR39]
gi|7190965|gb|AAF39725.1| hypothetical protein [Chlamydia pneumoniae phage CPAR39]
Length=553
Score = 296 bits (759), Expect = 6e-94, Method: Compositional matrix adjust.
Identities = 142/226 (63%), Positives = 166/226 (73%), Gaps = 10/226 (4%)
Query 2 FNVISPDARLQRPEYLGGTHSRVNVVPTAQTSSTDSVSPQSNLSAFGVLGDSAHGFNKSF 61
FNV SPDARLQR EYLGG+ + VN+ P QTSSTDS SPQ NL+A+G S F KSF
Sbjct 328 FNVQSPDARLQRAEYLGGSSTPVNISPIPQTSSTDSTSPQGNLAAYGTAIGSKRVFTKSF 387
Query 62 VEHGYVIGLCCLRADITYQQGLNRMWSRRQLFDFYWPTLAHLGEQVVYNKEIYAQGTA-- 119
EHG ++GL +RAD+ YQQGL+RMWSRR +DFYWP L+HLGEQ V NKEIY QG A
Sbjct 388 TEHGVILGLASVRADLNYQQGLDRMWSRRTRWDFYWPALSHLGEQAVLNKEIYCQGPAVK 447
Query 120 --------DDNGVFGYQERYAEYRYKPSMITGKLRSTDAQSLDVWHLAQRFDSLPKLNQD 171
D VFGYQER+AEYRYK S ITGK RS SLD WHLAQ+F++LP L+ +
Sbjct 448 DAQNGNVVVDEQVFGYQERFAEYRYKTSKITGKFRSNATGSLDAWHLAQQFENLPTLSPE 507
Query 172 FIEENPPINRVIAVQNEPQFFADFWFDLKTSRPMPVYSVPGLVDHF 217
FIEENPP++RV+AV EP F D WF L+ +RPMPVYSVPGL+DHF
Sbjct 508 FIEENPPMDRVVAVDTEPDFLLDGWFSLRCARPMPVYSVPGLIDHF 553
>gi|77020115|ref|YP_338238.1| putative major coat protein [Chlamydia phage 4]
gi|59940014|gb|AAX12543.1| putative major coat protein [Chlamydia phage 4]
Length=554
Score = 296 bits (757), Expect = 1e-93, Method: Compositional matrix adjust.
Identities = 142/226 (63%), Positives = 165/226 (73%), Gaps = 10/226 (4%)
Query 2 FNVISPDARLQRPEYLGGTHSRVNVVPTAQTSSTDSVSPQSNLSAFGVLGDSAHGFNKSF 61
FNV SPDARLQR EYLGG+ + VN+ P QTSSTDS SPQ NL+A+G S F KSF
Sbjct 329 FNVQSPDARLQRAEYLGGSSTPVNISPIPQTSSTDSTSPQGNLAAYGTAIGSKRVFTKSF 388
Query 62 VEHGYVIGLCCLRADITYQQGLNRMWSRRQLFDFYWPTLAHLGEQVVYNKEIYAQGTA-- 119
EHG ++GL +RAD+ YQQGL+RMWSRR +DFYWP L+HLGEQ V NKEIY QG A
Sbjct 389 TEHGVILGLASVRADLNYQQGLDRMWSRRTRWDFYWPALSHLGEQAVLNKEIYCQGPAVK 448
Query 120 --------DDNGVFGYQERYAEYRYKPSMITGKLRSTDAQSLDVWHLAQRFDSLPKLNQD 171
D VFGYQER+AEYRYK S ITGK RS SLD WHLAQ F++LP L+ +
Sbjct 449 DAQNGNVVVDEQVFGYQERFAEYRYKTSKITGKFRSNATSSLDSWHLAQEFENLPTLSPE 508
Query 172 FIEENPPINRVIAVQNEPQFFADFWFDLKTSRPMPVYSVPGLVDHF 217
FIEENPP++RV+AV EP F D WF L+ +RPMPVYSVPGL+DHF
Sbjct 509 FIEENPPMDRVLAVNTEPDFLLDGWFSLRCARPMPVYSVPGLIDHF 554
>gi|575096093|emb|CDL66973.1| unnamed protein product [uncultured bacterium]
Length=574
Score = 295 bits (756), Expect = 3e-93, Method: Compositional matrix adjust.
Identities = 134/216 (62%), Positives = 166/216 (77%), Gaps = 0/216 (0%)
Query 2 FNVISPDARLQRPEYLGGTHSRVNVVPTAQTSSTDSVSPQSNLSAFGVLGDSAHGFNKSF 61
F V +PD+RLQRPEYLGG S N+ P AQTSST+ +SPQ N++A+G+ G + FNKSF
Sbjct 359 FGVTNPDSRLQRPEYLGGRSSMFNINPVAQTSSTNDISPQGNMAAYGIHGRTYRAFNKSF 418
Query 62 VEHGYVIGLCCLRADITYQQGLNRMWSRRQLFDFYWPTLAHLGEQVVYNKEIYAQGTADD 121
E G VIGLC +RAD+TYQQG RMW R+ DFYWP AHLGEQ V N+EIY QGT+ D
Sbjct 419 TEFGVVIGLCSVRADLTYQQGTERMWFRKDDLDFYWPEFAHLGEQAVLNQEIYVQGTSAD 478
Query 122 NGVFGYQERYAEYRYKPSMITGKLRSTDAQSLDVWHLAQRFDSLPKLNQDFIEENPPINR 181
GVFGYQERYAEYRYKP+ ITG+ RST Q+LDVWHLAQ+FDSLPKL FI+++PP++R
Sbjct 479 TGVFGYQERYAEYRYKPNKITGQFRSTYKQTLDVWHLAQKFDSLPKLGDQFIQDHPPVSR 538
Query 182 VIAVQNEPQFFADFWFDLKTSRPMPVYSVPGLVDHF 217
V+AV + P F D F L+ RP+P++S+PGL+ HF
Sbjct 539 VVAVPSYPHFLLDVKFHLQCVRPLPLFSIPGLMPHF 574
>gi|9634949|ref|NP_054647.1| structural protein [Chlamydia phage 2]
gi|7406589|emb|CAB85589.1| structural protein [Chlamydia phage 2]
Length=565
Score = 295 bits (755), Expect = 3e-93, Method: Compositional matrix adjust.
Identities = 140/224 (63%), Positives = 164/224 (73%), Gaps = 8/224 (4%)
Query 2 FNVISPDARLQRPEYLGGTHSRVNVVPTAQTSSTDSVSPQSNLSAFGVLGDSAHGFNKSF 61
FNV SPDARLQR EYLGG+ + VN+ P QTSSTDS SPQ NL+A+G S F KSF
Sbjct 342 FNVQSPDARLQRAEYLGGSSTPVNISPIPQTSSTDSTSPQGNLAAYGTAIGSKRVFTKSF 401
Query 62 VEHGYVIGLCCLRADITYQQGLNRMWSRRQLFDFYWPTLAHLGEQVVYNKEIYAQGTADD 121
EHG ++GL +RAD+ YQQGL+RMWSRR +DFYWP L+HLGEQ V NKEIY QG +
Sbjct 402 TEHGVILGLASVRADLNYQQGLDRMWSRRTRWDFYWPALSHLGEQAVLNKEIYCQGPSVK 461
Query 122 NG--------VFGYQERYAEYRYKPSMITGKLRSTDAQSLDVWHLAQRFDSLPKLNQDFI 173
N VFGYQER+AEYRYK S ITGK RS SLD WHLAQ F++LP L+ +FI
Sbjct 462 NSGGEIVDDQVFGYQERFAEYRYKTSKITGKFRSNATSSLDSWHLAQEFENLPTLSPEFI 521
Query 174 EENPPINRVIAVQNEPQFFADFWFDLKTSRPMPVYSVPGLVDHF 217
EENPP++RV+AV EP F D WF L+ +RPMPVYSVPG +DHF
Sbjct 522 EENPPMDRVLAVSTEPDFLLDGWFSLRCARPMPVYSVPGFIDHF 565
Lambda K H a alpha
0.321 0.137 0.426 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 795278171475