bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-27_CDS_annotation_glimmer3.pl_2_5
Length=334
Score E
Sequences producing significant alignments: (Bits) Value
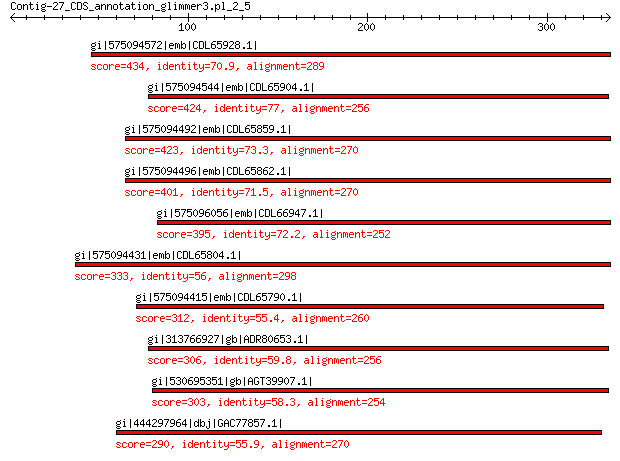
gi|575094572|emb|CDL65928.1| unnamed protein product 434 1e-145
gi|575094544|emb|CDL65904.1| unnamed protein product 424 2e-141
gi|575094492|emb|CDL65859.1| unnamed protein product 423 3e-141
gi|575094496|emb|CDL65862.1| unnamed protein product 401 2e-132
gi|575096056|emb|CDL66947.1| unnamed protein product 395 5e-130
gi|575094431|emb|CDL65804.1| unnamed protein product 333 4e-106
gi|575094415|emb|CDL65790.1| unnamed protein product 312 4e-98
gi|313766927|gb|ADR80653.1| putative major coat protein 306 6e-96
gi|530695351|gb|AGT39907.1| major capsid protein 303 1e-94
gi|444297964|dbj|GAC77857.1| major capsid protein 290 4e-93
>gi|575094572|emb|CDL65928.1| unnamed protein product [uncultured bacterium]
Length=556
Score = 434 bits (1116), Expect = 1e-145, Method: Compositional matrix adjust.
Identities = 205/291 (70%), Positives = 244/291 (84%), Gaps = 12/291 (4%)
Query 46 DVESVATGVG--FDAPTSRDGSMYLDNLWAIQSGNVTAATINQLRMAFQIQKLYEKDARG 103
+V S TG+G F PT N+WA++SG+V ATINQLR+AFQ+QKLYEKDARG
Sbjct 276 EVGSDGTGIGQNFWTPT---------NMWAVESGDVGMATINQLRLAFQLQKLYEKDARG 326
Query 104 GTRYIEILKSHFGVTSPDARLQRPEYLGGNRIPVNINQVVQSSATQSSGTPLGDTAAFSV 163
GTRY EI++SHFGV SPD+RLQRPEYLGGNRIP+N+NQ++Q S + + +PLG A SV
Sbjct 327 GTRYTEIIRSHFGVVSPDSRLQRPEYLGGNRIPINVNQIIQQSQS-TEQSPLGALAGMSV 385
Query 164 TTDVHGDFIKSFVEHGFVIGIMVARYDHTYQQGLERFWSRRDRLDYYFPVFANIGEQPIL 223
TTD + DFIKSFVEHG++IG++VARYDHTYQQGL+R WSR+DR D+Y+PV ANIGEQ +L
Sbjct 386 TTDKNSDFIKSFVEHGYIIGLVVARYDHTYQQGLDRMWSRKDRFDFYWPVLANIGEQAVL 445
Query 224 NKEIYAQGTVQDNEVFGYQEAWADYRYKPSRVAGEMRSKAPTSLDVWHLADEYTQLPKLS 283
NKEIY G+ D+EVFGYQEAWA+YRYKP+RV GEMRS AP SLDVWHL D+Y+ LP LS
Sbjct 446 NKEIYIDGSDTDDEVFGYQEAWAEYRYKPNRVCGEMRSSAPQSLDVWHLGDDYSSLPYLS 505
Query 284 DAWIREDKTNVDRVLAVTSAVSNQMFADLYIQCKATRPMPMYSIPGLIDHH 334
D+WIREDKTNVDRVLAVTS+VS+Q+FAD+YI KATRPMPMYSIPGLIDHH
Sbjct 506 DSWIREDKTNVDRVLAVTSSVSDQLFADIYICNKATRPMPMYSIPGLIDHH 556
>gi|575094544|emb|CDL65904.1| unnamed protein product [uncultured bacterium]
Length=551
Score = 424 bits (1089), Expect = 2e-141, Method: Compositional matrix adjust.
Identities = 197/256 (77%), Positives = 225/256 (88%), Gaps = 1/256 (0%)
Query 78 NVTAATINQLRMAFQIQKLYEKDARGGTRYIEILKSHFGVTSPDARLQRPEYLGGNRIPV 137
N TAA+INQLR+AFQIQ+LYE+DARGGTRYIEILKSHFGVTSPDARLQRPEYLGGNRIP+
Sbjct 296 NATAASINQLRLAFQIQRLYERDARGGTRYIEILKSHFGVTSPDARLQRPEYLGGNRIPI 355
Query 138 NINQVVQSSATQSSGTPLGDTAAFSVTTDVHGDFIKSFVEHGFVIGIMVARYDHTYQQGL 197
NINQV+Q S T S+ +P G+ S+TTD + DF+KSFVEHGFVIG+MVARYDHTYQQGL
Sbjct 356 NINQVLQQSETTST-SPQGNPVGQSLTTDTNADFVKSFVEHGFVIGLMVARYDHTYQQGL 414
Query 198 ERFWSRRDRLDYYFPVFANIGEQPILNKEIYAQGTVQDNEVFGYQEAWADYRYKPSRVAG 257
ERFWSR+DR DYY+PVFA+IGEQ +LNKEIY GT D+EVFGYQEA+ADYRYKPSRV G
Sbjct 415 ERFWSRKDRFDYYWPVFAHIGEQAVLNKEIYTSGTAVDDEVFGYQEAYADYRYKPSRVTG 474
Query 258 EMRSKAPTSLDVWHLADEYTQLPKLSDAWIREDKTNVDRVLAVTSAVSNQMFADLYIQCK 317
EMRS AP SLDVWHLAD+Y LP LSD+WIRE + VDRVLAV+S VS Q+F D+YIQ +
Sbjct 475 EMRSAAPQSLDVWHLADDYASLPSLSDSWIRESASTVDRVLAVSSNVSAQLFCDIYIQNR 534
Query 318 ATRPMPMYSIPGLIDH 333
+TRPMPMYS+PGLIDH
Sbjct 535 STRPMPMYSVPGLIDH 550
>gi|575094492|emb|CDL65859.1| unnamed protein product [uncultured bacterium]
Length=551
Score = 423 bits (1087), Expect = 3e-141, Method: Compositional matrix adjust.
Identities = 198/272 (73%), Positives = 231/272 (85%), Gaps = 4/272 (1%)
Query 65 SMYLDNLWAIQSG--NVTAATINQLRMAFQIQKLYEKDARGGTRYIEILKSHFGVTSPDA 122
SM NLWA S ++ ATINQLR AFQIQKLYE+DARGGTRYIEILKSHFGVTSPDA
Sbjct 282 SMIPTNLWADLSTATDLPVATINQLRTAFQIQKLYERDARGGTRYIEILKSHFGVTSPDA 341
Query 123 RLQRPEYLGGNRIPVNINQVVQSSATQSSGTPLGDTAAFSVTTDVHGDFIKSFVEHGFVI 182
RLQRPEYLGG+R+P+NINQV+QSS T TP G+ AA+S+TTD H +F KSFVEHGF+I
Sbjct 342 RLQRPEYLGGSRVPININQVIQSSET--GATPQGNAAAYSLTTDSHSEFTKSFVEHGFII 399
Query 183 GIMVARYDHTYQQGLERFWSRRDRLDYYFPVFANIGEQPILNKEIYAQGTVQDNEVFGYQ 242
G+MVARYDH+YQQGL+RFWSR+DR DYY+PVFAN+GE + NKEI+AQGT D+EVFGYQ
Sbjct 400 GLMVARYDHSYQQGLQRFWSRKDRFDYYWPVFANLGEMAVKNKEIFAQGTDVDDEVFGYQ 459
Query 243 EAWADYRYKPSRVAGEMRSKAPTSLDVWHLADEYTQLPKLSDAWIREDKTNVDRVLAVTS 302
EAWADYRYKPS V GEMRS+ SLD+WHLAD+Y LP LSD+WIRED + V+RVLAV+
Sbjct 460 EAWADYRYKPSVVTGEMRSQYAQSLDIWHLADDYENLPSLSDSWIREDSSTVNRVLAVSD 519
Query 303 AVSNQMFADLYIQCKATRPMPMYSIPGLIDHH 334
+VS Q+F D+YI+C ATRPMP+YSIPGLIDHH
Sbjct 520 SVSAQLFCDIYIRCLATRPMPLYSIPGLIDHH 551
>gi|575094496|emb|CDL65862.1| unnamed protein product [uncultured bacterium]
Length=568
Score = 401 bits (1030), Expect = 2e-132, Method: Compositional matrix adjust.
Identities = 193/270 (71%), Positives = 222/270 (82%), Gaps = 4/270 (1%)
Query 65 SMYLDNLWAIQSGNVTAATINQLRMAFQIQKLYEKDARGGTRYIEILKSHFGVTSPDARL 124
S+Y DNL+A SG TA TINQLRMAFQIQKLYEKDAR G+RY E+++SHF VT DAR+
Sbjct 303 SVYPDNLYA-SSG--TATTINQLRMAFQIQKLYEKDARAGSRYRELIRSHFSVTPLDARM 359
Query 125 QRPEYLGGNRIPVNINQVVQSSATQSSGTPLGDTAAFSVTTDVHGDFIKSFVEHGFVIGI 184
Q PEYLGGNRIP+NINQVVQ+S T S +P G+ A S+T+D HGDFIKSF EHG +IG+
Sbjct 360 QVPEYLGGNRIPININQVVQTSQT-SDVSPQGNVAGQSLTSDSHGDFIKSFTEHGMLIGV 418
Query 185 MVARYDHTYQQGLERFWSRRDRLDYYFPVFANIGEQPILNKEIYAQGTVQDNEVFGYQEA 244
VARYDHTYQQG+ + WSR+ R DYY+PV ANIGEQ +LNKEIYAQGT QD EVFGYQEA
Sbjct 419 AVARYDHTYQQGVSKLWSRKTRFDYYWPVLANIGEQAVLNKEIYAQGTAQDEEVFGYQEA 478
Query 245 WADYRYKPSRVAGEMRSKAPTSLDVWHLADEYTQLPKLSDAWIREDKTNVDRVLAVTSAV 304
WA+YRYKPS V GEMRS A TSLD WH AD+Y LPKLS WI+EDKTN+DRVLAV+S+V
Sbjct 479 WAEYRYKPSIVTGEMRSSARTSLDSWHFADDYNSLPKLSADWIKEDKTNIDRVLAVSSSV 538
Query 305 SNQMFADLYIQCKATRPMPMYSIPGLIDHH 334
SNQ FAD YI+ + TR +P YSIPGLIDHH
Sbjct 539 SNQYFADFYIENETTRALPFYSIPGLIDHH 568
>gi|575096056|emb|CDL66947.1| unnamed protein product [uncultured bacterium]
Length=570
Score = 395 bits (1014), Expect = 5e-130, Method: Compositional matrix adjust.
Identities = 182/253 (72%), Positives = 207/253 (82%), Gaps = 1/253 (0%)
Query 83 TINQLRMAFQIQKLYEKDARGGTRYIEILKSHFGVTSPDARLQRPEYLGGNRIPVNINQV 142
TINQLRMAFQIQK YEK ARGG+RY E+++S FGVTSPDARLQR EYLGGNRIP+NINQV
Sbjct 318 TINQLRMAFQIQKFYEKQARGGSRYTEVIRSFFGVTSPDARLQRSEYLGGNRIPININQV 377
Query 143 VQSSATQS-SGTPLGDTAAFSVTTDVHGDFIKSFVEHGFVIGIMVARYDHTYQQGLERFW 201
+Q S T S S TP G S TTD H DF KSF EHGF+IG+M ARYDHTYQQG++R W
Sbjct 378 IQQSGTGSASTTPQGTVVGMSQTTDTHSDFTKSFTEHGFIIGVMCARYDHTYQQGIDRMW 437
Query 202 SRRDRLDYYFPVFANIGEQPILNKEIYAQGTVQDNEVFGYQEAWADYRYKPSRVAGEMRS 261
SR+D+ DYY+PVF+NIGEQ I NKEIYAQG D+EVFGYQEAWA+YRYKPSRV GEMRS
Sbjct 438 SRKDKFDYYWPVFSNIGEQAIKNKEIYAQGNATDDEVFGYQEAWAEYRYKPSRVTGEMRS 497
Query 262 KAPTSLDVWHLADEYTQLPKLSDAWIREDKTNVDRVLAVTSAVSNQMFADLYIQCKATRP 321
SLDVWHLAD+Y++LP LSD WIRED ++RVLAV+ SNQ FAD+Y++ TRP
Sbjct 498 SYAQSLDVWHLADDYSKLPSLSDEWIREDAKTLNRVLAVSDQNSNQFFADIYVKNLCTRP 557
Query 322 MPMYSIPGLIDHH 334
MPMYSIPGLIDHH
Sbjct 558 MPMYSIPGLIDHH 570
>gi|575094431|emb|CDL65804.1| unnamed protein product [uncultured bacterium]
Length=560
Score = 333 bits (853), Expect = 4e-106, Method: Compositional matrix adjust.
Identities = 167/298 (56%), Positives = 202/298 (68%), Gaps = 17/298 (6%)
Query 37 NFGSNLGGTDVESVATGVGFDAPTSRDGSMYLDNLWAIQSGNVTAATINQLRMAFQIQKL 96
NF + GG+ ES A Y NLWA S AAT+NQLR AFQ+QKL
Sbjct 280 NFETKAGGSFSESGAVAA------------YPTNLWA--SPVTAAATVNQLRQAFQVQKL 325
Query 97 YEKDARGGTRYIEILKSHFGVTSPDARLQRPEYLGGNRIPVNINQVVQSSATQSSGTPLG 156
EKDARGGTRY EILK+HFGVT+ DAR+Q PEYLGG ++P+N++QVVQ+SA+ + +P G
Sbjct 326 LEKDARGGTRYREILKNHFGVTTSDARMQIPEYLGGCKVPINVSQVVQTSAS-TDASPQG 384
Query 157 DTAAFSVTTDVHGDFIKSFVEHGFVIGIMVARYDHTYQQGLERFWSRRDRLDYYFPVFAN 216
+TAA SVT F KSF EHGF+IG+ AR +YQQG+ER WSR+DRLDYYFPV AN
Sbjct 385 NTAAISVTPFSKSMFTKSFDEHGFIIGVATARTAQSYQQGIERMWSRKDRLDYYFPVLAN 444
Query 217 IGEQPILNKEIYAQGTVQDNEVFGYQEAWADYRYKPSRVAGEMRSKAPTSLDVWHLADEY 276
IGEQ ILNKEIYAQG +D+E FGYQEAWADYRYKP+ + G RS A SLD WH +Y
Sbjct 445 IGEQAILNKEIYAQGNAKDDEAFGYQEAWADYRYKPNTICGRFRSNAQQSLDAWHYGQDY 504
Query 277 TQLPKLSDAWIREDKTNVDRVLAVTSAVSNQMFADLYIQCKATRPMPMYSIPGLIDHH 334
+LP LS W+ + + R LAV + A+ CK R MP+YSIPGLIDH+
Sbjct 505 DKLPTLSTDWMEQSDIEMKRTLAVQT--EPDFIANFRFNCKTVRVMPLYSIPGLIDHN 560
>gi|575094415|emb|CDL65790.1| unnamed protein product [uncultured bacterium]
Length=569
Score = 312 bits (800), Expect = 4e-98, Method: Compositional matrix adjust.
Identities = 144/260 (55%), Positives = 188/260 (72%), Gaps = 1/260 (0%)
Query 71 LWAIQSGNVTAATINQLRMAFQIQKLYEKDARGGTRYIEILKSHFGVTSPDARLQRPEYL 130
L A+ + +IN LR A +Q + E DARGGTRY+EILK+ FGV+SPDARLQR EY+
Sbjct 308 LTAVAENSTNFLSINDLRQAIALQHILEADARGGTRYVEILKNEFGVSSPDARLQRSEYI 367
Query 131 GGNRIPVNINQVVQSSATQSSGTPLGDTAAFSVTTDVHGDFIKSFVEHGFVIGIMVARYD 190
GG RIP+N++QV+QSSA+ ++ +P G+ AA+S+TT + S VEHG+++G+ R D
Sbjct 368 GGERIPINVSQVIQSSASDTT-SPQGNAAAYSLTTSANTIRAYSAVEHGYILGLAAIRVD 426
Query 191 HTYQQGLERFWSRRDRLDYYFPVFANIGEQPILNKEIYAQGTVQDNEVFGYQEAWADYRY 250
H+YQQGL R W+R DR YY P+ AN+GEQ +LN+EIYAQGT D EVFGYQEAWADYRY
Sbjct 427 HSYQQGLSRMWTRSDRFSYYHPMLANLGEQAVLNQEIYAQGTTADTEVFGYQEAWADYRY 486
Query 251 KPSRVAGEMRSKAPTSLDVWHLADEYTQLPKLSDAWIREDKTNVDRVLAVTSAVSNQMFA 310
+ + + GEMRS SLD WH D+YT LP+LS+ WI+E + N+DR LAV S S+Q
Sbjct 487 RTNMITGEMRSTYAQSLDAWHYGDKYTDLPRLSNDWIKEGQENIDRTLAVQSENSHQFIC 546
Query 311 DLYIQCKATRPMPMYSIPGL 330
+LY RPMP+YS+PGL
Sbjct 547 NLYFDQTWVRPMPIYSVPGL 566
>gi|313766927|gb|ADR80653.1| putative major coat protein [Uncultured Microviridae]
Length=533
Score = 306 bits (783), Expect = 6e-96, Method: Compositional matrix adjust.
Identities = 153/256 (60%), Positives = 188/256 (73%), Gaps = 5/256 (2%)
Query 78 NVTAATINQLRMAFQIQKLYEKDARGGTRYIEILKSHFGVTSPDARLQRPEYLGGNRIPV 137
N TAATINQLR AFQIQ+LYEKDARGGTRY EIL+SHFGVTSPDARLQRPEYLGG + V
Sbjct 267 NATAATINQLREAFQIQRLYEKDARGGTRYTEILQSHFGVTSPDARLQRPEYLGGQKTEV 326
Query 138 NINQVVQSSATQSSGTPLGDTAAFSVTTDVHGDFIKSFVEHGFVIGIMVARYDHTYQQGL 197
+ V Q+S+T S+ +P G+ AA T G F KSFVEHG +IG+ D TYQQG+
Sbjct 327 MMQTVPQTSSTDST-SPQGNLAALGTATS-RGGFSKSFVEHGVLIGLACVFADLTYQQGM 384
Query 198 ERFWSRRDRLDYYFPVFANIGEQPILNKEIYAQGTVQDNEVFGYQEAWADYRYKPSRVAG 257
R WSRRDR D+Y+P A++GEQ +LN+EIY QGT D + FGYQE +A+YRYKPS++ G
Sbjct 385 NRMWSRRDRWDFYWPSLAHLGEQAVLNQEIYTQGTSADTQTFGYQERFAEYRYKPSQITG 444
Query 258 EMRSKAPTSLDVWHLADEYTQLPKLSDAWIREDKTNVDRVLAVTSAVSNQMFADLYIQCK 317
+MRS A +LD WHLA ++T LP L+ ++I E+ VDRV+AV S + D Y K
Sbjct 445 KMRSNATGTLDAWHLAQDFTALPALNASFIEENPP-VDRVIAVPS--EPEFIWDWYFDLK 501
Query 318 ATRPMPMYSIPGLIDH 333
TRPMP+YS+PGLIDH
Sbjct 502 TTRPMPVYSVPGLIDH 517
>gi|530695351|gb|AGT39907.1| major capsid protein [Marine gokushovirus]
Length=539
Score = 303 bits (775), Expect = 1e-94, Method: Compositional matrix adjust.
Identities = 148/257 (58%), Positives = 181/257 (70%), Gaps = 4/257 (2%)
Query 80 TAATINQLRMAFQIQKLYEKDARGGTRYIEILKSHFGVTSPDARLQRPEYLGGNRIPVNI 139
TAATIN +R +FQIQ+L E+DARGGTRY EI++SHFGV SPDAR+QRPEYLGG P+ +
Sbjct 283 TAATINAIRQSFQIQRLLERDARGGTRYTEIVRSHFGVISPDARMQRPEYLGGGSAPIIV 342
Query 140 NQVVQSSATQSSGT--PLGDTAAFSVTTDVHGDFIKSFVEHGFVIGIMVARYDHTYQQGL 197
N V Q SA+ +SGT PLG A F SF EHG V+G+ R D TYQQGL
Sbjct 343 NPVAQQSASGASGTDTPLGTLGAVGTGLASGHGFASSFTEHGVVVGLCSVRADLTYQQGL 402
Query 198 ERFWSRRDRLDYYFPVFANIGEQPILNKEIYAQGTVQDNEVFGYQEAWADYRYKPSRVAG 257
R +SR R D++FPVF+++GEQPILNKE+YA GT D++VFGYQEAWA+YRYKPS+V G
Sbjct 403 HRMFSRSTRYDFFFPVFSHLGEQPILNKELYATGTSTDDDVFGYQEAWAEYRYKPSQVTG 462
Query 258 EMRSKAPTSLDVWHLADEYTQLPKLSDAWIREDKTNVDRVLAVTSAVSNQMFA-DLYIQC 316
MRS A +LD WHLA + LP L+ +I ED VDRV+AV S + Q F D +
Sbjct 463 LMRSTAAGTLDAWHLAQNFGSLPTLNSTFI-EDTPPVDRVVAVGSEANGQQFIFDAFFDI 521
Query 317 KATRPMPMYSIPGLIDH 333
RPMPMYS+PGL+DH
Sbjct 522 NMARPMPMYSVPGLVDH 538
>gi|444297964|dbj|GAC77857.1| major capsid protein, partial [uncultured marine virus]
Length=285
Score = 290 bits (743), Expect = 4e-93, Method: Compositional matrix adjust.
Identities = 151/275 (55%), Positives = 194/275 (71%), Gaps = 15/275 (5%)
Query 60 TSRDG-SMYLDNLWAIQSGNVTAATINQLRMAFQIQKLYEKDARGGTRYIEILKSHFGVT 118
TS +G S+Y D + TA+TINQLR AFQIQKL E+DARGGTRYIEI+KSHFGVT
Sbjct 21 TSTEGQSLYAD------LSDATASTINQLRQAFQIQKLLERDARGGTRYIEIVKSHFGVT 74
Query 119 SPDARLQRPEYLGGNRIPVNINQVVQSSATQSSGTP--LGDTAAFSVTTDVHGDFIKSFV 176
SPD R RPEYLGG PVNIN VV ++A S+G+P LGD AA++ T + F KSF
Sbjct 75 SPDLRATRPEYLGGGSNPVNINPVV-NTAGFSAGSPQNLGDLAAYATTVIQNNGFTKSFT 133
Query 177 EHGFVIGIMVARYDHTYQQGLERFWSRRDRLDYYFPVFANIGEQPILNKEIYAQGTV--Q 234
EH +IG++ R D TYQQG+ R WSR DR D+YFP A+IGEQ +LNKEIYA G Q
Sbjct 134 EHCIIIGLVSVRADLTYQQGMNRMWSRSDRYDFYFPALAHIGEQAVLNKEIYAIGNQADQ 193
Query 235 DNEVFGYQEAWADYRYKPSRVAGEMRSKAPTSLDVWHLADEYTQLPKLSDAWIREDKTNV 294
D +VFGYQE +A+YRYKPS++ G+ RS A ++LD WHL+ ++T LP+L+ A+I+E+ +
Sbjct 194 DEDVFGYQERFAEYRYKPSQITGKFRSTAASTLDAWHLSQKFTSLPELNSAFIQENPP-M 252
Query 295 DRVLAVTSAVSNQMFADLYIQCKATRPMPMYSIPG 329
DRV+AV S + D Y + + RPMP+Y +P
Sbjct 253 DRVVAVDS--EPEFIWDSYFKMRCVRPMPLYGVPA 285
Lambda K H a alpha
0.318 0.133 0.404 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1917593351550