bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-27_CDS_annotation_glimmer3.pl_2_4
Length=250
Score E
Sequences producing significant alignments: (Bits) Value
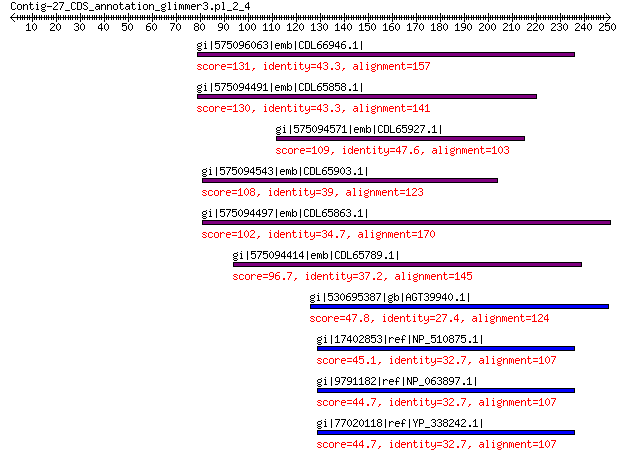
gi|575096063|emb|CDL66946.1| unnamed protein product 131 4e-34
gi|575094491|emb|CDL65858.1| unnamed protein product 130 8e-34
gi|575094571|emb|CDL65927.1| unnamed protein product 109 3e-26
gi|575094543|emb|CDL65903.1| unnamed protein product 108 1e-25
gi|575094497|emb|CDL65863.1| unnamed protein product 102 4e-23
gi|575094414|emb|CDL65789.1| unnamed protein product 96.7 6e-21
gi|530695387|gb|AGT39940.1| portal protein 47.8 0.001
gi|17402853|ref|NP_510875.1| capsid protein VP3 45.1 0.007
gi|9791182|ref|NP_063897.1| capsid protein VP3 44.7 0.010
gi|77020118|ref|YP_338242.1| putative capsid protein 44.7 0.011
>gi|575096063|emb|CDL66946.1| unnamed protein product [uncultured bacterium]
Length=163
Score = 131 bits (330), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 68/157 (43%), Positives = 92/157 (59%), Gaps = 0/157 (0%)
Query 79 MPMFETWHREQQHFCSEPGCGEKILYSPEFDRFGVMTLKESGKEDLYAFIQSHKDSVDLH 138
M F+T + +E G E I Y +D G + L+E+G+ +LY IQSH +SVDLH
Sbjct 1 MMEFKTQYDPHDRIFTEAGSREHITYGGHYDDEGRVVLEETGRINLYDEIQSHAESVDLH 60
Query 139 KIMDRFNAGDASALQKVQGMFGDFSEMPQTYAGLLNHMIEAEQTFMSLPLETREKFGQSF 198
+M R++ GD L + QG FGD + PQTYA LNHM E E+ FMSLPL+ R KFG SF
Sbjct 61 VLMQRYSCGDVDCLSQRQGFFGDVLDFPQTYAEALNHMQEMERQFMSLPLDVRAKFGHSF 120
Query 199 HAWLAQAGSVSWLEAMGMVTPPTSQNSAGEPPAAPQS 235
+LA +G +LE +G P S + PA P++
Sbjct 121 SEFLAASGDDDFLERLGFKPEPESAPAVDPVPAVPEA 157
>gi|575094491|emb|CDL65858.1| unnamed protein product [uncultured bacterium]
Length=176
Score = 130 bits (328), Expect = 8e-34, Method: Compositional matrix adjust.
Identities = 61/141 (43%), Positives = 87/141 (62%), Gaps = 0/141 (0%)
Query 79 MPMFETWHREQQHFCSEPGCGEKILYSPEFDRFGVMTLKESGKEDLYAFIQSHKDSVDLH 138
M F+T PG K++YSP +D GV+ L+ +G+E+LY FIQSH S D+H
Sbjct 1 MVKFKTQFHSHARVFQRPGDPIKVVYSPRYDENGVLDLQPTGEENLYDFIQSHAQSTDIH 60
Query 139 KIMDRFNAGDASALQKVQGMFGDFSEMPQTYAGLLNHMIEAEQTFMSLPLETREKFGQSF 198
I+DRF +G+ L ++QG + D S+MP+TYA +LN +I EQTF LP+E ++KFG SF
Sbjct 61 VILDRFASGETDVLSQIQGFYADASDMPKTYAEVLNAVIAGEQTFDRLPVEIKQKFGNSF 120
Query 199 HAWLAQAGSVSWLEAMGMVTP 219
WL+ + + E MG P
Sbjct 121 STWLSSMDNPDFAERMGFPKP 141
>gi|575094571|emb|CDL65927.1| unnamed protein product [uncultured bacterium]
Length=133
Score = 109 bits (273), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 49/103 (48%), Positives = 68/103 (66%), Gaps = 0/103 (0%)
Query 112 GVMTLKESGKEDLYAFIQSHKDSVDLHKIMDRFNAGDASALQKVQGMFGDFSEMPQTYAG 171
G + L E+GK++LY IQSHKDSVDL+ ++ RFN G+ L ++QG + D S MP+TYA
Sbjct 8 GAIDLVENGKKNLYDEIQSHKDSVDLNLLLQRFNNGEVDVLSRMQGTYADLSNMPKTYAD 67
Query 172 LLNHMIEAEQTFMSLPLETREKFGQSFHAWLAQAGSVSWLEAM 214
+LN + + E F+SLP++ R KF SF WL GS W+ M
Sbjct 68 MLNLIKKGEADFLSLPVDVRAKFDHSFEKWLVTFGSQDWIVNM 110
>gi|575094543|emb|CDL65903.1| unnamed protein product [uncultured bacterium]
Length=145
Score = 108 bits (269), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 48/123 (39%), Positives = 74/123 (60%), Gaps = 0/123 (0%)
Query 81 MFETWHREQQHFCSEPGCGEKILYSPEFDRFGVMTLKESGKEDLYAFIQSHKDSVDLHKI 140
MF + + G I+Y +D GV L SG++++Y IQSH+DSVD+H +
Sbjct 1 MFRSQFDDHDRVYCSSGSPIHIVYQGRYDDDGVFDLFPSGQDNIYDQIQSHRDSVDIHVL 60
Query 141 MDRFNAGDASALQKVQGMFGDFSEMPQTYAGLLNHMIEAEQTFMSLPLETREKFGQSFHA 200
+DR+ GD L QG++GDF+ MP +Y+ +LN ++ E+ FM LP++ R K+G SF
Sbjct 61 LDRYQRGDVDVLSARQGVYGDFTGMPASYSEILNAVLAGERAFMDLPVDERAKYGHSFAQ 120
Query 201 WLA 203
WL+
Sbjct 121 WLS 123
>gi|575094497|emb|CDL65863.1| unnamed protein product [uncultured bacterium]
Length=170
Score = 102 bits (254), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 59/171 (35%), Positives = 94/171 (55%), Gaps = 5/171 (3%)
Query 81 MFET-WHREQQHFCSEPGCGEKILYSPEFDRFGVMTLKESGKEDLYAFIQSHKDSVDLHK 139
+F T ++R ++ F S G G + Y G L++ G+ DLYA IQS+KDS D++
Sbjct 2 LFRTKYNRNKEVFKSTSGDGYQPTYKMRVHEDGKRELEKVGRIDLYAQIQSYKDSCDINY 61
Query 140 IMDRFNAGDASALQKVQGMFGDFSEMPQTYAGLLNHMIEAEQTFMSLPLETREKFGQSFH 199
I++RF GD SAL K+QG++GDF+ MP A L +++AE F +LP++ R +F S
Sbjct 62 ILERFARGDESALSKIQGVYGDFTAMPTNLAELQQRVVDAEALFYNLPVDIRAEFNHSPS 121
Query 200 AWLAQAGSVSWLEAMGMVTPPTSQNSAGEPPAAPQSPEGGETDPAPASPAG 250
+ + G+ + +A+G +TP +P +P + DP P G
Sbjct 122 EFYSAIGTDKFNKAVG-ITPQVDPQPIIDPQPVISTP---QVDPQPVVQKG 168
>gi|575094414|emb|CDL65789.1| unnamed protein product [uncultured bacterium]
Length=167
Score = 96.7 bits (239), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 54/149 (36%), Positives = 79/149 (53%), Gaps = 5/149 (3%)
Query 94 SEPGCGEKILYSPEFDRFGVMTLKESGKEDLYAFIQSHKDSVDLHKIMDRFNAGDASALQ 153
+EPG + Y + G L++ G+ + YA IQS+KD D+H I+ R+ AGD S L
Sbjct 18 NEPGNELEPHYIEKIGENGRTYLEKDGETNTYAEIQSYKDECDVHSILCRYFAGDTSVLS 77
Query 154 KVQGMFGDFSEMPQTYAGLLNHMIEAEQTFMSLPLETREKFGQSFHAWLAQAGSVSWLEA 213
+ QG++ D +++P TY + N M E F LPLE + KF SF+ W + AGS W +
Sbjct 78 R-QGVYIDATQLPTTYHEMYNLMAEQRDKFDQLPLEIKRKFDNSFNVWASTAGSEEWYKL 136
Query 214 MGMVTPPTSQ----NSAGEPPAAPQSPEG 238
MG+V + N +GE EG
Sbjct 137 MGIVQNAENNKPEGNGSGENEGENAQKEG 165
>gi|530695387|gb|AGT39940.1| portal protein [Marine gokushovirus]
Length=164
Score = 47.8 bits (112), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 34/129 (26%), Positives = 63/129 (49%), Gaps = 10/129 (8%)
Query 126 AFIQSH-KDSVDLHKIMDRFN-AGDASALQKVQGMFGDFSEMPQTYAGLLNHMIEAEQTF 183
+ Q H KD ++ I+ + + G +Q+ Q +GDFS++ Y L+ + +A+Q F
Sbjct 32 SLTQQHFKDECEVINIIKKHDRNGIIEHVQRGQARYGDFSQVAD-YREALDLVRDAQQEF 90
Query 184 MSLPLETREKFGQS---FHAWLAQAGSVSWLEAMGMVTPPTSQNSAGEPPAAPQSPEGGE 240
MS+P + R+KF F+ +++ + L+ MG + P G+P + P
Sbjct 91 MSVPSDIRKKFDNDPGKFYEFVSNPDNKEELKQMGFIETP----EVGKPSSVPTKALSEA 146
Query 241 TDPAPASPA 249
+P+ A A
Sbjct 147 GEPSTAQEA 155
>gi|17402853|ref|NP_510875.1| capsid protein VP3 [Guinea pig Chlamydia phage]
Length=148
Score = 45.1 bits (105), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 35/126 (28%), Positives = 58/126 (46%), Gaps = 20/126 (16%)
Query 129 QSHKDSVDLHKIMDRFNA-GDASALQKVQGMFGDFSEMPQTYAGLLNHMIEAEQTFMSLP 187
Q +KD D++ I+ + NA G +++ + D + P Y+ LN +IEA++ F SLP
Sbjct 24 QHNKDECDINNIVAKLNATGVLEHVERRSPRYMDCMD-PMEYSEALNVVIEAQEQFDSLP 82
Query 188 LETREKFGQSFHA---WLAQAGSVSWLEAMGMV---------------TPPTSQNSAGEP 229
+ RE+FG A +L++ + +A+G V P QN A +
Sbjct 83 AKVRERFGNDPEAMLDFLSREENYEEAKALGFVYEDGTSGAPHTFCEADPKDDQNVANQE 142
Query 230 PAAPQS 235
P Q
Sbjct 143 PGLAQK 148
>gi|9791182|ref|NP_063897.1| capsid protein VP3 [Chlamydia pneumoniae phage CPAR39]
gi|7190961|gb|AAF39721.1| capsid protein VP3 [Chlamydia pneumoniae phage CPAR39]
Length=148
Score = 44.7 bits (104), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 35/126 (28%), Positives = 58/126 (46%), Gaps = 20/126 (16%)
Query 129 QSHKDSVDLHKIMDRFNA-GDASALQKVQGMFGDFSEMPQTYAGLLNHMIEAEQTFMSLP 187
Q +KD D++ I+ + NA G +++ + D + P Y+ LN +IEA++ F SLP
Sbjct 24 QHNKDECDINNIVAKLNATGVLEHVERRSPRYMDCMD-PMEYSEALNVVIEAQEQFDSLP 82
Query 188 LETREKFGQSFHA---WLAQAGSVSWLEAMGMV---------------TPPTSQNSAGEP 229
+ RE+FG A +L++ + +A+G V P QN A +
Sbjct 83 AKVRERFGNDPEAMLDFLSREENYEEAKALGFVYEDGTSGAPQTFFEADPKDDQNVANQE 142
Query 230 PAAPQS 235
P Q
Sbjct 143 PGLAQK 148
>gi|77020118|ref|YP_338242.1| putative capsid protein [Chlamydia phage 4]
gi|59940017|gb|AAX12546.1| putative capsid protein [Chlamydia phage 4]
Length=148
Score = 44.7 bits (104), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 35/126 (28%), Positives = 58/126 (46%), Gaps = 20/126 (16%)
Query 129 QSHKDSVDLHKIMDRFNA-GDASALQKVQGMFGDFSEMPQTYAGLLNHMIEAEQTFMSLP 187
Q +KD D++ I+ + NA G +++ + D + P Y+ LN +IEA++ F SLP
Sbjct 24 QHNKDECDINNIVAKLNATGVLEHVERRSPRYMDCMD-PMEYSEALNVVIEAQEQFDSLP 82
Query 188 LETREKFGQSFHA---WLAQAGSVSWLEAMGMV---------------TPPTSQNSAGEP 229
+ RE+FG A +L++ + +A+G V P QN A +
Sbjct 83 AKIRERFGNDPEAMLDFLSREENYEEAKALGFVYEDGTSGAPQTFFEADPKDDQNVANQE 142
Query 230 PAAPQS 235
P Q
Sbjct 143 PGLAQK 148
Lambda K H a alpha
0.319 0.132 0.411 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1113602771862