bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-26_CDS_annotation_glimmer3.pl_2_4
Length=255
Score E
Sequences producing significant alignments: (Bits) Value
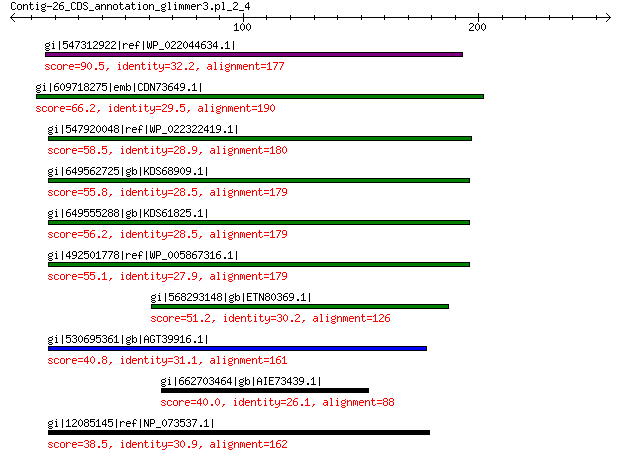
gi|547312922|ref|WP_022044634.1| putative replication initiation... 90.5 5e-18
gi|609718275|emb|CDN73649.1| conserved hypothetical protein 66.2 1e-09
gi|547920048|ref|WP_022322419.1| putative replication protein 58.5 7e-07
gi|649562725|gb|KDS68909.1| hypothetical protein M096_3339 55.8 5e-06
gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 56.2 5e-06
gi|492501778|ref|WP_005867316.1| hypothetical protein 55.1 1e-05
gi|568293148|gb|ETN80369.1| hypothetical protein NECAME_18023 51.2 3e-04
gi|530695361|gb|AGT39916.1| replication initiator 40.8 0.51
gi|662703464|gb|AIE73439.1| Transcriptional regulator PchR 40.0 1.1
gi|12085145|ref|NP_073537.1| putative replication initiation pro... 38.5 2.9
>gi|547312922|ref|WP_022044634.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
gi|524208442|emb|CCZ76638.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
Length=320
Score = 90.5 bits (223), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 57/193 (30%), Positives = 103/193 (53%), Gaps = 30/193 (16%)
Query 16 IIFATLTFSEESLKKLEYDEKEPNKAPQKAISLFRKRWWKKYKTPLKHWLITEMGHDNTK 75
+F TLTF+++SL+K D KA+ LF R+ K Y ++HW + E G +
Sbjct 74 CLFVTLTFNDDSLEKFSKDT-------NKAVRLFLDRFRKVYGKQIRHWFVCEFGTLH-G 125
Query 76 RIHLHGIIWT--ELTEEQFEKE-----------WGYGWIFFGYEVSEKTINYIVKYVTKR 122
R H HGI++ + + ++ + W YG++F GY VS++T +YI KYVTK
Sbjct 126 RPHYHGILFNVPQALIDGYDSDMPGHHPLLASCWKYGFVFVGY-VSDETCSYITKYVTKS 184
Query 123 ---DEANPEFNGKIFTSKGIGIGYINKNSLNKHRYQDKFTEETYRAASGIKLALPTYYKQ 179
D+ P ++ +S GIG Y+N + H+ ++ + + +G + A+P YY
Sbjct 185 INGDKVRP----RVISSFGIGSNYLNTEESSLHKLGNQ-RYQPFMVLNGFQQAMPRYYYN 239
Query 180 KLWTDQERESLRI 192
K+++D +++++ +
Sbjct 240 KIFSDVDKQNMVV 252
>gi|609718275|emb|CDN73649.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=265
Score = 66.2 bits (160), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 56/198 (28%), Positives = 95/198 (48%), Gaps = 16/198 (8%)
Query 12 LSKNIIFATLTFSEESLKKLEYDEKEPNKAPQKAISLFRKRWWKKYKTPLKHWLITEMGH 71
+SK+ F TLT+S+ L Y + + LF KR K K+ +K++L+ E G
Sbjct 49 VSKSAHFVTLTYSDV---YLPYSDNGLISLDYRDFQLFMKRARKLQKSKIKYFLVGEYGA 105
Query 72 DNTKRIHLHGIIWTELTEEQFEKEWGYGWIFFGYEVSEKTINYIVKYVTKR-------DE 124
T R H H I++ + F EW G + G V+ K+I Y +KY TK D
Sbjct 106 -QTYRPHYHAIVFGVENIDAFLGEWRMGNVHAG-TVTAKSIYYTLKYCTKSITEGPDKDP 163
Query 125 ANPEFNGKIFTSKGIGIGYINKNSLNKHRYQDKFTEETYRAASGIKLALPTYYKQKLWTD 184
+ K SKG+G+ ++ ++ + +Y ++ G +ALP YY+ K+++D
Sbjct 164 DDDRKPEKALMSKGLGLSHLTESMI---KYYKDDVSRSFSLLGGTTIALPRYYRDKVFSD 220
Query 185 QER-ESLRIIKEEQQVKY 201
E+ L I + +++Y
Sbjct 221 IEKVHRLVSITDYLEIRY 238
>gi|547920048|ref|WP_022322419.1| putative replication protein [Parabacteroides merdae CAG:48]
gi|524592960|emb|CDD13572.1| putative replication protein [Parabacteroides merdae CAG:48]
Length=278
Score = 58.5 bits (140), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 52/199 (26%), Positives = 102/199 (51%), Gaps = 25/199 (13%)
Query 17 IFATLTFSEESL--KKLEYDEKEPNKA--PQKAISLFRKRWWKKYKT-PLKHWLITEMGH 71
+F TLT+ +E L +++ D + N A ++ + LF KR KKY+ +++++ +E G
Sbjct 41 LFVTLTYDDEHLPIERIGSDLFQTNVAVVSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGA 100
Query 72 DNTKRIHLHGIIWT-----ELTEEQFEKEWGYGWIFFGYEVSEKTINYIVKYVTKRDEAN 126
N R H H I++ ++ + + W G++ + ++ K I Y+ KY+ ++
Sbjct 101 KNG-RPHYHMILFGFPFTGKMAGDLLAECWQNGFVQ-AHPLTIKEIAYVCKYMYEKSMC- 157
Query 127 PE-------FNGKIFTSK--GIGIGYINKNSLNKHRYQDKFTEETYRAASGIKLALPTYY 177
PE + + S+ GIG G++ + + +R + + RA +G K+A+P YY
Sbjct 158 PEILRDEKKYKPFMLCSRNPGIGFGFMKADIIEFYR---RHPRDYVRAWAGHKMAMPRYY 214
Query 178 KQKLWTDQERESLRIIKEE 196
KL+ D + L+ ++EE
Sbjct 215 ADKLYDDDMKAFLKEMREE 233
>gi|649562725|gb|KDS68909.1| hypothetical protein M096_3339 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=250
Score = 55.8 bits (133), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 51/197 (26%), Positives = 96/197 (49%), Gaps = 23/197 (12%)
Query 17 IFATLTFSEESLKKLEYDEK----EPNKAPQKAISLFRKRWWKKY-KTPLKHWLITEMGH 71
+F TLT+ +E + E ++ I LF KR KKY + L+++L +E G
Sbjct 12 LFVTLTYDDEHIPTAMIGEDLFKTTVGVVSKRDIQLFMKRLRKKYAQYRLRYFLTSEYG- 70
Query 72 DNTKRIHLHGIIWT-----ELTEEQFEKEWGYGWIFFGYEVSEKTINYIVKYVTKRD--- 123
R H H I++ + + + W G++ + ++ K I+Y+ KY+ ++
Sbjct 71 SQGGRPHYHMILFGFPFTGKHGGDLLAECWKNGFVQ-AHPLTTKEISYVTKYMYEKSMIP 129
Query 124 ---EANPEFNGKIFTSKGIGIGY--INKNSLNKHRYQDKFTEETYRAASGIKLALPTYYK 178
+ E+ + SK GIGY + + L+ +R + + RA +G+++A+P YY
Sbjct 130 DILKGVKEYQPFMLCSKMPGIGYHFLREQILDFYRLHPR---DYVRAFNGMRMAMPRYYA 186
Query 179 QKLWTDQERESLRIIKE 195
KL+ D +E L+ ++E
Sbjct 187 DKLYDDDMKEYLKELRE 203
>gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649560564|gb|KDS66872.1| hypothetical protein M095_2449 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561011|gb|KDS67298.1| hypothetical protein M095_2409 [Parabacteroides distasonis str.
3999B T(B) 4]
Length=284
Score = 56.2 bits (134), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 51/197 (26%), Positives = 96/197 (49%), Gaps = 23/197 (12%)
Query 17 IFATLTFSEESLKKLEYDEK----EPNKAPQKAISLFRKRWWKKY-KTPLKHWLITEMGH 71
+F TLT+ +E + E ++ I LF KR KKY + L+++L +E G
Sbjct 46 LFVTLTYDDEHIPTAMIGEDLFKTTVGVVSKRDIQLFMKRLRKKYAQYRLRYFLTSEYG- 104
Query 72 DNTKRIHLHGIIWT-----ELTEEQFEKEWGYGWIFFGYEVSEKTINYIVKYVTKRD--- 123
R H H I++ + + + W G++ + ++ K I+Y+ KY+ ++
Sbjct 105 SQGGRPHYHMILFGFPFTGKHGGDLLAECWKNGFVQ-AHPLTTKEISYVTKYMYEKSMIP 163
Query 124 ---EANPEFNGKIFTSKGIGIGY--INKNSLNKHRYQDKFTEETYRAASGIKLALPTYYK 178
+ E+ + SK GIGY + + L+ +R + + RA +G+++A+P YY
Sbjct 164 DILKGVKEYQPFMLCSKMPGIGYHFLREQILDFYRLHPR---DYVRAFNGMRMAMPRYYA 220
Query 179 QKLWTDQERESLRIIKE 195
KL+ D +E L+ ++E
Sbjct 221 DKLYDDDMKEYLKELRE 237
>gi|492501778|ref|WP_005867316.1| hypothetical protein [Parabacteroides distasonis]
gi|409230407|gb|EKN23271.1| hypothetical protein HMPREF1059_03256 [Parabacteroides distasonis
CL09T03C24]
Length=284
Score = 55.1 bits (131), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 50/197 (25%), Positives = 94/197 (48%), Gaps = 23/197 (12%)
Query 17 IFATLTFSEESLKKLEYDE----KEPNKAPQKAISLFRKRWWKKY-KTPLKHWLITEMGH 71
+F TLT+ +E + E ++ I LF KR KKY + L+++L +E G
Sbjct 46 LFVTLTYDDEHMPTAMIGEDLFKSTVGVVSKRDIQLFMKRLRKKYDQYRLRYFLTSEYG- 104
Query 72 DNTKRIHLHGIIWT-----ELTEEQFEKEWGYGWIFFGYEVSEKTINYIVKYVTKRDEAN 126
R H H I++ + + + W G++ + ++ K I Y+ KY+ ++
Sbjct 105 SQGGRPHYHMILFGFPFTGKHGGDLLAECWKNGFVQ-AHPLTTKEIAYVTKYMYEKSMVP 163
Query 127 P------EFNGKIFTSKGIGIGY--INKNSLNKHRYQDKFTEETYRAASGIKLALPTYYK 178
E+ + S+ GIGY + + L+ +R + + RA +G+++A+P YY
Sbjct 164 DILKDVKEYQPFMLCSRIPGIGYHFLREQILDFYRLHPR---DYVRAFNGMRMAMPRYYA 220
Query 179 QKLWTDQERESLRIIKE 195
KL+ D +E L+ ++E
Sbjct 221 DKLYDDDMKEYLKELRE 237
>gi|568293148|gb|ETN80369.1| hypothetical protein NECAME_18023 [Necator americanus]
Length=345
Score = 51.2 bits (121), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 38/138 (28%), Positives = 63/138 (46%), Gaps = 18/138 (13%)
Query 61 LKHWLITEMGHDNTKRIHLHGIIWTELTEEQFEKEWGYGWIFFGYEVSE----KTINYIV 116
LK+++ E G R H H II+ + F W G V K+I Y +
Sbjct 93 LKYYMCGEYGSQRF-RPHYHAIIFGVPQDSLFADAWTLNGDSLGGVVVGTVTGKSIAYTM 151
Query 117 KYVTK--------RDEANPEFNGKIFTSKGIGIGYINKNSLNKHRYQDKFTEETYRAASG 168
KY+ K RD+ PEF+ SKG+G+ Y+ + H+ + + G
Sbjct 152 KYIDKSTWKQKHGRDDRVPEFS---LMSKGMGVSYLTPQMVEYHK--EDISRLFCTREGG 206
Query 169 IKLALPTYYKQKLWTDQE 186
++A+P YY+QK+++D +
Sbjct 207 SRIAMPRYYRQKIYSDDD 224
>gi|530695361|gb|AGT39916.1| replication initiator [Marine gokushovirus]
Length=289
Score = 40.8 bits (94), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 50/198 (25%), Positives = 70/198 (35%), Gaps = 50/198 (25%)
Query 17 IFATLTFSEESLKKLEYDEKEPNKAPQKAISLFRKRWWKKYKTPLKHWLITEMGHDNTKR 76
F TLTF E + K K P F KR KKY ++ + E G D KR
Sbjct 61 CFITLTFDNEHIAK----RKNPESLDNTEFQRFMKRLRKKYPHKIRFFHCGEYG-DQNKR 115
Query 77 IHLHGIIWTE----------------LTEEQFEKEWGYGWIFFGYEVSEKTINYIVKYVT 120
H H +++ ++ + W YG+ G VS T Y +YV
Sbjct 116 PHYHALLFGHDFKDKKLWSNKGDFKLFVSQELAELWPYGFHTIG-AVSFDTAAYCARYVM 174
Query 121 KR---------------------DEANPEFNGKIFTSKGIGIGYINKNSLNKHRYQDKFT 159
K+ +E PE+ S+ GIGY K+ Y D
Sbjct 175 KKVTGDAAASHYREVDLETGEVINEIKPEY---CTMSRMPGIGY---EWYQKYGYHDCHK 228
Query 160 EETYRAASGIKLALPTYY 177
+ Y +G K+ P YY
Sbjct 229 HD-YIVINGYKVRPPRYY 245
>gi|662703464|gb|AIE73439.1| Transcriptional regulator PchR [Synechocystis sp. PCC 6714]
Length=329
Score = 40.0 bits (92), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 23/92 (25%), Positives = 40/92 (43%), Gaps = 9/92 (10%)
Query 65 LITEMGHDNTKRIHLHGIIWTELTEEQFEKEWGYGWIFFGYEVSEKTINY--IVKYVTKR 122
L T +GH +T+R L IW +L +++F + W E + + ++
Sbjct 40 LPTYLGHGHTRRFELSSGIWLDLIDKEFTQPWA-----LKMPAHEHLVQFTILLSGAVDY 94
Query 123 DEANPEFNGKI--FTSKGIGIGYINKNSLNKH 152
DE P K+ F+ GI GY+ + +H
Sbjct 95 DETYPTLGAKMGYFSGSGISPGYVARYGRLRH 126
>gi|12085145|ref|NP_073537.1| putative replication initiation protein [Bdellovibrio phage phiMH2K]
gi|75089164|sp|Q9G050.1|REP_BPPHM RecName: Full=Replication-associated protein VP4; Short=Rep;
Short=VP4 [Bdellovibrio phage phiMH2K]
gi|12017993|gb|AAG45349.1|AF306496_10 Vp4 [Bdellovibrio phage phiMH2K]
Length=315
Score = 38.5 bits (88), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 50/195 (26%), Positives = 89/195 (46%), Gaps = 51/195 (26%)
Query 17 IFATLTFSEESLKK-----LEYDE--KEPNKAPQKAISLFRKRWWKKYKTPLKHWLITEM 69
IF TLT+ +E LK +++D K N+ + +S K+ + PL + + E
Sbjct 64 IFLTLTYDDEHLKSDRLQWIDFDLFIKRLNEKLNRGLS-------KENRRPLPYMVTGEY 116
Query 70 GHDNTKRIHLHGIIW------------TELTEEQFEKE-----WGYGWIFFGYEVSEKTI 112
G D TKR H H +I+ TEL E+ + E W +G I FG V+ +
Sbjct 117 G-DKTKRPHWHVLIFNFRPDDAKKHYVTELGEQVYTSEFIRDLWTHGNIEFG-SVTLDSA 174
Query 113 NYIVKYVTKR----DEANPEFNGKIFTSKGIGIGYINKNSLNKHRYQDKFTEETYR---- 164
+Y+ +Y K+ ++ + +++ TSK IG ++ +K+ E+T+
Sbjct 175 SYVARYAAKKLAHGNDQDHDYHPIHNTSKKHAIG---------KKWIEKYHEQTFSRGYV 225
Query 165 -AASGIKLALPTYYK 178
+G + +P YY+
Sbjct 226 VLPNGSQGPIPRYYQ 240
Lambda K H a alpha
0.315 0.132 0.398 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1166131204497