bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-25_CDS_annotation_glimmer3.pl_2_4
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
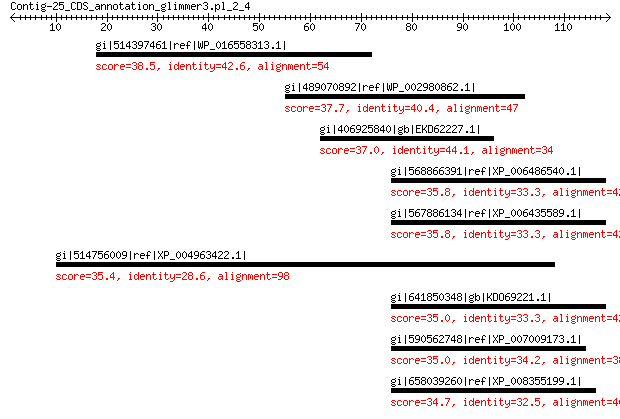
gi|514397461|ref|WP_016558313.1| hypothetical protein 38.5 0.35
gi|489070892|ref|WP_002980862.1| conserved hypothetical protein 37.7 0.88
gi|406925840|gb|EKD62227.1| Endonuclease/exonuclease/phosphatase 37.0 1.2
gi|568866391|ref|XP_006486540.1| PREDICTED: BEL1-like homeodomai... 35.8 4.2
gi|567886134|ref|XP_006435589.1| hypothetical protein CICLE_v100... 35.8 4.3
gi|514756009|ref|XP_004963422.1| PREDICTED: uncharacterized prot... 35.4 5.8
gi|641850348|gb|KDO69221.1| hypothetical protein CISIN_1g040555mg 35.0 6.1
gi|590562748|ref|XP_007009173.1| POX family protein, putative 35.0 7.6
gi|658039260|ref|XP_008355199.1| PREDICTED: BEL1-like homeodomai... 34.7 8.7
>gi|514397461|ref|WP_016558313.1| hypothetical protein [Rhizobium grahamii]
gi|512773237|gb|EPE94053.1| hypothetical protein RGCCGE502_32097 [Rhizobium grahamii CCGE
502]
Length=183
Score = 38.5 bits (88), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 23/55 (42%), Positives = 29/55 (53%), Gaps = 1/55 (2%)
Query 18 QDFKAYKNRGGIGDRAENLRELIGLERVVRQDFGYEG-TYYVTEDGETYDEEYVD 71
+D NR I + +LR+LIG VVRQ G G TY VTE G ++Y D
Sbjct 80 KDLARLANREDIPNIQYSLRKLIGAGLVVRQGSGRSGVTYLVTESGRAVTDQYAD 134
>gi|489070892|ref|WP_002980862.1| conserved hypothetical protein [Chryseobacterium gleum]
gi|300504506|gb|EFK35646.1| PGAP1-like protein [Chryseobacterium gleum ATCC 35910]
Length=608
Score = 37.7 bits (86), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 24/47 (51%), Gaps = 5/47 (11%)
Query 55 TYYVTEDGETYDEEYVDAHNPRKTGVKFYEVSNWRYNRKKGIYEPVI 101
TYY ETY Y+D H P KFY + W Y R G++ P+I
Sbjct 190 TYYC----ETYFRPYMDNH-PLNQPQKFYSLGAWGYTRLTGLFSPII 231
>gi|406925840|gb|EKD62227.1| Endonuclease/exonuclease/phosphatase, partial [uncultured bacterium]
Length=241
Score = 37.0 bits (84), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 23/34 (68%), Gaps = 3/34 (9%)
Query 62 GETYDEEYVDAHNPRKTGVKFYEVSNWRYNRKKG 95
G+T DEE+V+ +NP T ++++NWR RK G
Sbjct 10 GDTADEEFVELYNPTDTP---FDLTNWRLARKTG 40
>gi|568866391|ref|XP_006486540.1| PREDICTED: BEL1-like homeodomain protein 9-like [Citrus sinensis]
Length=395
Score = 35.8 bits (81), Expect = 4.2, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 76 RKTGVKFYEVSNWRYNRKKGIYEPVIRRIVMIEIKEIQLTLE 117
++TG+ +VSNW N + +++P++ + M+EI + Q T E
Sbjct 232 KQTGLSKNQVSNWFINARVRLWKPMVEEVHMLEIGQTQTTAE 273
>gi|567886134|ref|XP_006435589.1| hypothetical protein CICLE_v10033544mg, partial [Citrus clementina]
gi|557537785|gb|ESR48829.1| hypothetical protein CICLE_v10033544mg, partial [Citrus clementina]
Length=485
Score = 35.8 bits (81), Expect = 4.3, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 76 RKTGVKFYEVSNWRYNRKKGIYEPVIRRIVMIEIKEIQLTLE 117
++TG+ +VSNW N + +++P++ + M+EI + Q T E
Sbjct 351 KQTGLSKNQVSNWFINARVRLWKPMVEEVHMLEIGQTQTTAE 392
>gi|514756009|ref|XP_004963422.1| PREDICTED: uncharacterized protein LOC101769462 [Setaria italica]
Length=353
Score = 35.4 bits (80), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 28/104 (27%), Positives = 47/104 (45%), Gaps = 8/104 (8%)
Query 10 DRRHLNAKQDF---KAYKNRGGIGDRAENLRELIGLERVVRQDF-GYEGTYYVTEDGETY 65
D H+ AK F Y+NR G ++N+ ++ + G+EG YY+ + G
Sbjct 175 DGTHIEAKIKFDEQTPYRNRHGFP--SQNVMAVVSFDMTFTHVVAGWEGKYYLVDSGYAN 232
Query 66 DEEYVDAHNPRKTGVKFYEVSNWRYNRKKGIYEP--VIRRIVMI 107
++V + + + + SNW Y K GI P +IVM+
Sbjct 233 TLKFVAPYRGDRYHIGSFRGSNWHYKVKGGIPYPYETQVKIVMV 276
>gi|641850348|gb|KDO69221.1| hypothetical protein CISIN_1g040555mg, partial [Citrus sinensis]
Length=313
Score = 35.0 bits (79), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 76 RKTGVKFYEVSNWRYNRKKGIYEPVIRRIVMIEIKEIQLTLE 117
++TG+ +VSNW N + +++P++ + M+EI + Q T E
Sbjct 153 KQTGLSKNQVSNWFINARVRLWKPMVEEVHMLEIGQTQTTSE 194
>gi|590562748|ref|XP_007009173.1| POX family protein, putative [Theobroma cacao]
gi|508726086|gb|EOY17983.1| POX family protein, putative [Theobroma cacao]
Length=503
Score = 35.0 bits (79), Expect = 7.6, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 26/38 (68%), Gaps = 0/38 (0%)
Query 76 RKTGVKFYEVSNWRYNRKKGIYEPVIRRIVMIEIKEIQ 113
R+TG+ +VSNW N + +++P++ I M+E+++ Q
Sbjct 338 RQTGLSRTQVSNWFINARVRLWKPMVEEIHMLELRQSQ 375
>gi|658039260|ref|XP_008355199.1| PREDICTED: BEL1-like homeodomain protein 9 [Malus domestica]
Length=311
Score = 34.7 bits (78), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 13/40 (33%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 76 RKTGVKFYEVSNWRYNRKKGIYEPVIRRIVMIEIKEIQLT 115
++TG+ +VSNW N + +++P++ I M+E ++ Q T
Sbjct 123 KQTGLSRSQVSNWFINARVRLWKPMVEEIHMLETRQAQKT 162
Lambda K H a alpha
0.318 0.139 0.406 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 442112487102