bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-24_CDS_annotation_glimmer3.pl_2_5
Length=421
Score E
Sequences producing significant alignments: (Bits) Value
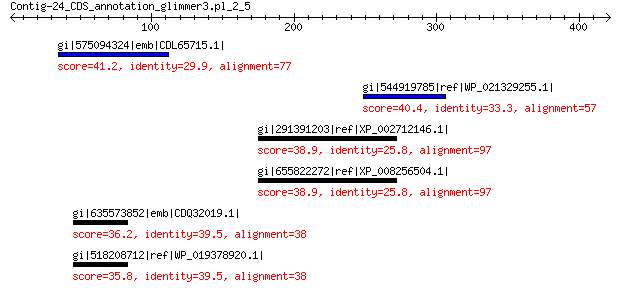
gi|575094324|emb|CDL65715.1| unnamed protein product 41.2 1.0
gi|544919785|ref|WP_021329255.1| hypothetical protein 40.4 3.3
gi|291391203|ref|XP_002712146.1| PREDICTED: alpha-aminoadipic se... 38.9 8.3
gi|655822272|ref|XP_008256504.1| PREDICTED: alpha-aminoadipic se... 38.9 9.3
gi|635573852|emb|CDQ32019.1| GIY-YIG nuclease superfamily protein 36.2 9.6
gi|518208712|ref|WP_019378920.1| hypothetical protein 35.8 9.8
>gi|575094324|emb|CDL65715.1| unnamed protein product [uncultured bacterium]
Length=370
Score = 41.2 bits (95), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 23/77 (30%), Positives = 41/77 (53%), Gaps = 0/77 (0%)
Query 35 FGGISARRNWKYKQKEMALQQQYALEQMSKSAEFQLAHDKQMFDYQNSYNDPAAVLERNF 94
F GI A++ + K + + A K AE+Q+ +++ + +YN+P+AV++R
Sbjct 18 FSGIGAKKRQEAAIKAQREENEKARNWQQKMAEWQVGIERENLADERAYNNPSAVMKRLK 77
Query 95 SAGLNPAAVLGQSGVGV 111
AGLNP + G G+
Sbjct 78 DAGLNPDLMYGSGASGL 94
>gi|544919785|ref|WP_021329255.1| hypothetical protein [Treponema socranskii]
gi|538293860|gb|ERF61708.1| hypothetical protein HMPREF1325_1344 [Treponema socranskii subsp.
socranskii VPI DR56BR1116 = ATCC 35536]
gi|544005847|gb|ERK05105.1| hypothetical protein HMPREF0860_1608 [Treponema socranskii subsp.
socranskii VPI DR56BR1116 = ATCC 35536]
Length=1341
Score = 40.4 bits (93), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 35/59 (59%), Gaps = 2/59 (3%)
Query 249 KARTGYIDELI--EKELQLLTVRAIYLKSSASNQEQLARVNELTADDLENWFDVNWNTQ 305
+++TG++ E I E E +L T + +KS+ SNQ++L ++N +T W V W+ +
Sbjct 383 ESKTGFVPEKIIGETEAKLSTTIGLKIKSTTSNQDKLKKINVITCGTARTWNGVQWSAE 441
>gi|291391203|ref|XP_002712146.1| PREDICTED: alpha-aminoadipic semialdehyde synthase, mitochondrial
isoform X1 [Oryctolagus cuniculus]
gi|655822155|ref|XP_008256501.1| PREDICTED: alpha-aminoadipic semialdehyde synthase, mitochondrial
isoform X1 [Oryctolagus cuniculus]
gi|655822219|ref|XP_008256502.1| PREDICTED: alpha-aminoadipic semialdehyde synthase, mitochondrial
isoform X1 [Oryctolagus cuniculus]
gi|655822247|ref|XP_008256503.1| PREDICTED: alpha-aminoadipic semialdehyde synthase, mitochondrial
isoform X1 [Oryctolagus cuniculus]
Length=926
Score = 38.9 bits (89), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 25/97 (26%), Positives = 48/97 (49%), Gaps = 1/97 (1%)
Query 175 RSQTLDKDLRERLMKAQAGLAEAGITESASRASLNAAITLSYSIDNELKDAAFGYNLEMI 234
R+Q+L D +++++ +G + E SR S N IT+ + N+++ + YN+ +
Sbjct 471 RAQSLSMDTKKKVLVLGSGYVSEPVLEYLSRDS-NIEITVGSDMKNQIEQLSKKYNITPV 529
Query 235 KANLGKAKEEYYQLKARTGYIDELIEKELQLLTVRAI 271
N+GK +E+ L A + L+ L L +A
Sbjct 530 SMNIGKQEEKLDSLVATQDLVISLLPYVLHPLVAKAC 566
>gi|655822272|ref|XP_008256504.1| PREDICTED: alpha-aminoadipic semialdehyde synthase, mitochondrial
isoform X2 [Oryctolagus cuniculus]
Length=885
Score = 38.9 bits (89), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 25/97 (26%), Positives = 48/97 (49%), Gaps = 1/97 (1%)
Query 175 RSQTLDKDLRERLMKAQAGLAEAGITESASRASLNAAITLSYSIDNELKDAAFGYNLEMI 234
R+Q+L D +++++ +G + E SR S N IT+ + N+++ + YN+ +
Sbjct 430 RAQSLSMDTKKKVLVLGSGYVSEPVLEYLSRDS-NIEITVGSDMKNQIEQLSKKYNITPV 488
Query 235 KANLGKAKEEYYQLKARTGYIDELIEKELQLLTVRAI 271
N+GK +E+ L A + L+ L L +A
Sbjct 489 SMNIGKQEEKLDSLVATQDLVISLLPYVLHPLVAKAC 525
>gi|635573852|emb|CDQ32019.1| GIY-YIG nuclease superfamily protein [Virgibacillus halodenitrificans]
Length=97
Score = 36.2 bits (82), Expect = 9.6, Method: Composition-based stats.
Identities = 15/40 (38%), Positives = 30/40 (75%), Gaps = 2/40 (5%)
Query 45 KYKQKEMALQQQYALEQMSKSAEFQLAHD--KQMFDYQNS 82
K+ K+ A+Q++Y ++Q+S++ +FQL D K++ +Y+NS
Sbjct 55 KFPTKQAAMQKEYQIKQLSRTEKFQLIRDRLKEVIEYENS 94
>gi|518208712|ref|WP_019378920.1| hypothetical protein [Virgibacillus halodenitrificans]
Length=97
Score = 35.8 bits (81), Expect = 9.8, Method: Composition-based stats.
Identities = 15/40 (38%), Positives = 30/40 (75%), Gaps = 2/40 (5%)
Query 45 KYKQKEMALQQQYALEQMSKSAEFQLAHD--KQMFDYQNS 82
K+ K+ A+Q++Y ++Q+S++ +FQL D K++ +Y+NS
Sbjct 55 KFPTKQAAMQKEYQIKQLSRTEKFQLIRDRLKEVIEYENS 94
Lambda K H a alpha
0.314 0.128 0.357 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2743904169720