bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-24_CDS_annotation_glimmer3.pl_2_3
Length=97
Score E
Sequences producing significant alignments: (Bits) Value
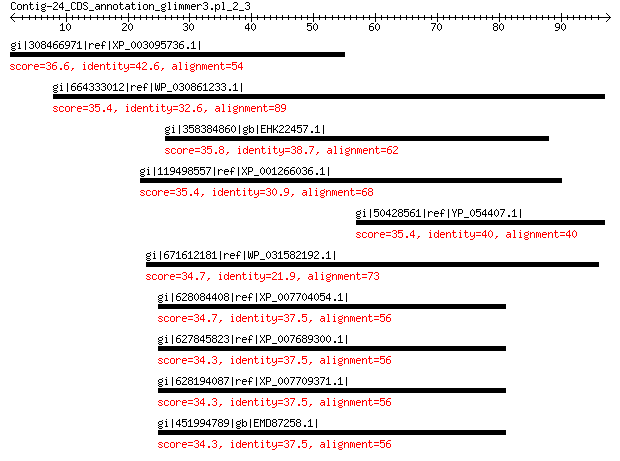
gi|308466971|ref|XP_003095736.1| hypothetical protein CRE_10575 36.6 1.4
gi|664333012|ref|WP_030861233.1| chemotaxis protein CheY 35.4 2.6
gi|358384860|gb|EHK22457.1| hypothetical protein TRIVIDRAFT_84034 35.8 2.7
gi|119498557|ref|XP_001266036.1| SET and WW domain protein 35.4 3.9
gi|50428561|ref|YP_054407.1| replication-associated protein 35.4 4.1
gi|671612181|ref|WP_031582192.1| hypothetical protein 34.7 5.7
gi|628084408|ref|XP_007704054.1| hypothetical protein COCSADRAFT... 34.7 7.1
gi|627845823|ref|XP_007689300.1| hypothetical protein COCMIDRAFT... 34.3 7.5
gi|628194087|ref|XP_007709371.1| hypothetical protein COCCADRAFT... 34.3 7.8
gi|451994789|gb|EMD87258.1| hypothetical protein COCHEDRAFT_1113604 34.3 8.4
>gi|308466971|ref|XP_003095736.1| hypothetical protein CRE_10575 [Caenorhabditis remanei]
gi|308244501|gb|EFO88453.1| hypothetical protein CRE_10575 [Caenorhabditis remanei]
Length=502
Score = 36.6 bits (83), Expect = 1.4, Method: Composition-based stats.
Identities = 23/58 (40%), Positives = 35/58 (60%), Gaps = 5/58 (9%)
Query 1 MLEYMIEDLPEYRSRGERIMSVLNGSGSVEVLP--GRP--DVQASDSDFRKGEDYDPP 54
ML+Y++E EYR +GE+IM L G + VLP G+ + S ++ KG+DY+ P
Sbjct 59 MLDYIVETANEYRGKGEKIMR-LQFIGKLLVLPLDGKTAQTMLQSTTELDKGDDYEFP 115
>gi|664333012|ref|WP_030861233.1| chemotaxis protein CheY [Streptomyces sp. NRRL F-2747]
Length=216
Score = 35.4 bits (80), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 29/89 (33%), Positives = 40/89 (45%), Gaps = 9/89 (10%)
Query 8 DLPEYRSRGERIMSVLNGSGSVEVLPGRPDVQASDSDFRKGEDYDPPLDFDPNSFSRIDK 67
D P YR E I+S L GSG +EV+ D + + R+ LD+ R+ +
Sbjct 16 DHPVYR---EGIVSALTGSGRIEVVAAVGDGRTALEAIREHAPAVALLDY------RMPE 66
Query 68 FDGLESGQGVIDDFLERQRSSSSAKSEKD 96
DGLE V D L + SA +E D
Sbjct 67 LDGLEVAHAVARDELPTRVLLLSATTESD 95
>gi|358384860|gb|EHK22457.1| hypothetical protein TRIVIDRAFT_84034 [Trichoderma virens Gv29-8]
Length=794
Score = 35.8 bits (81), Expect = 2.7, Method: Composition-based stats.
Identities = 24/67 (36%), Positives = 36/67 (54%), Gaps = 10/67 (15%)
Query 26 SGSVEVLPGRPDVQASDSDFRKGEDYDPPLDFDPNSFSRIDKFD-GLESG----QGVIDD 80
S V+VLPG +V++ +FRK +DP LD D ++D D L SG + ID+
Sbjct 18 SSGVDVLPGAGNVESELRNFRKQHRWDPFLDID-----KLDNIDEALASGNAEKEAAIDE 72
Query 81 FLERQRS 87
L ++ S
Sbjct 73 SLIQEDS 79
>gi|119498557|ref|XP_001266036.1| SET and WW domain protein [Neosartorya fischeri NRRL 181]
gi|119414200|gb|EAW24139.1| SET and WW domain protein [Neosartorya fischeri NRRL 181]
Length=967
Score = 35.4 bits (80), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 21/68 (31%), Positives = 39/68 (57%), Gaps = 4/68 (6%)
Query 22 VLNGSGSVEVLPGRPDVQASDSDFRKGEDYDPPLDFDPNSFSRIDKFDGLESGQGVIDDF 81
+ NG +VE + DV+ SD + K E+ D +D DP + + D ++ G+G ++
Sbjct 800 MTNGHNTVEDV----DVKLSDGEAEKVEENDTFMDSDPQGILKRKREDEMDIGEGAAEEA 855
Query 82 LERQRSSS 89
++RQ+SS+
Sbjct 856 MKRQKSST 863
>gi|50428561|ref|YP_054407.1| replication-associated protein [Opuntia virus X]
gi|38146353|gb|AAR11547.1| replication-associated protein [Opuntia virus X]
Length=1555
Score = 35.4 bits (80), Expect = 4.1, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 57 FDPNSFSRIDKFDGLESGQGVIDDFLERQRSSSSAKSEKD 96
+DPNS++ +DK D L + Q DFL++QR+ + + KD
Sbjct 435 WDPNSYNPLDKVDELSTEQLKAWDFLQKQRNPTQEEFHKD 474
>gi|671612181|ref|WP_031582192.1| hypothetical protein [Lachnospiraceae bacterium AC2028]
Length=582
Score = 34.7 bits (78), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 16/73 (22%), Positives = 38/73 (52%), Gaps = 0/73 (0%)
Query 23 LNGSGSVEVLPGRPDVQASDSDFRKGEDYDPPLDFDPNSFSRIDKFDGLESGQGVIDDFL 82
+N +++++ P++Q ++DF+K D + P + D + +I+ +E +F+
Sbjct 507 VNDISNLQLIEAMPNIQKQNTDFKKWYDEEYPTEKDKIQYRKINYIPDMEYDYSDFKEFI 566
Query 83 ERQRSSSSAKSEK 95
E++R+ EK
Sbjct 567 EKRRTLLKKAFEK 579
>gi|628084408|ref|XP_007704054.1| hypothetical protein COCSADRAFT_40358 [Bipolaris sorokiniana
ND90Pr]
gi|451846573|gb|EMD59882.1| hypothetical protein COCSADRAFT_40358 [Bipolaris sorokiniana
ND90Pr]
Length=388
Score = 34.7 bits (78), Expect = 7.1, Method: Composition-based stats.
Identities = 21/56 (38%), Positives = 25/56 (45%), Gaps = 0/56 (0%)
Query 25 GSGSVEVLPGRPDVQASDSDFRKGEDYDPPLDFDPNSFSRIDKFDGLESGQGVIDD 80
S SV P RP DSD+ DY PPLD P S+I K + + DD
Sbjct 250 ASVSVSPKPARPATNRDDSDYNAIPDYSPPLDTLPKGNSKILKAEWKGQALSLADD 305
>gi|627845823|ref|XP_007689300.1| hypothetical protein COCMIDRAFT_6491 [Bipolaris oryzae ATCC 44560]
gi|576930591|gb|EUC44169.1| hypothetical protein COCMIDRAFT_6491 [Bipolaris oryzae ATCC 44560]
Length=389
Score = 34.3 bits (77), Expect = 7.5, Method: Composition-based stats.
Identities = 21/56 (38%), Positives = 25/56 (45%), Gaps = 0/56 (0%)
Query 25 GSGSVEVLPGRPDVQASDSDFRKGEDYDPPLDFDPNSFSRIDKFDGLESGQGVIDD 80
S SV P RP DSD+ DY PPLD P S+I K + + DD
Sbjct 251 ASVSVSPKPARPATNRDDSDYNAIPDYSPPLDTLPKGNSKILKAEWKGQALSLADD 306
>gi|628194087|ref|XP_007709371.1| hypothetical protein COCCADRAFT_23883 [Bipolaris zeicola 26-R-13]
gi|576922160|gb|EUC36291.1| hypothetical protein COCCADRAFT_23883 [Bipolaris zeicola 26-R-13]
gi|578485038|gb|EUN22544.1| hypothetical protein COCVIDRAFT_41654 [Bipolaris victoriae FI3]
Length=392
Score = 34.3 bits (77), Expect = 7.8, Method: Composition-based stats.
Identities = 21/56 (38%), Positives = 25/56 (45%), Gaps = 0/56 (0%)
Query 25 GSGSVEVLPGRPDVQASDSDFRKGEDYDPPLDFDPNSFSRIDKFDGLESGQGVIDD 80
S SV P RP DSD+ DY PPLD P S+I K + + DD
Sbjct 254 ASVSVSPKPARPATNRDDSDYNAIPDYSPPLDTLPKGNSKILKAEWKGQALSLADD 309
>gi|451994789|gb|EMD87258.1| hypothetical protein COCHEDRAFT_1113604 [Bipolaris maydis C5]
gi|477583247|gb|ENI00347.1| hypothetical protein COCC4DRAFT_150697 [Bipolaris maydis ATCC
48331]
Length=388
Score = 34.3 bits (77), Expect = 8.4, Method: Composition-based stats.
Identities = 21/56 (38%), Positives = 25/56 (45%), Gaps = 0/56 (0%)
Query 25 GSGSVEVLPGRPDVQASDSDFRKGEDYDPPLDFDPNSFSRIDKFDGLESGQGVIDD 80
S SV P RP DSD+ DY PPLD P S+I K + + DD
Sbjct 250 ASVSVSPKPARPATNRDDSDYNAIPDYSPPLDTLPKGNSKILKAEWKGQALSLADD 305
Lambda K H a alpha
0.311 0.135 0.378 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 428386497840