bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-20_CDS_annotation_glimmer3.pl_2_9
Length=334
Score E
Sequences producing significant alignments: (Bits) Value
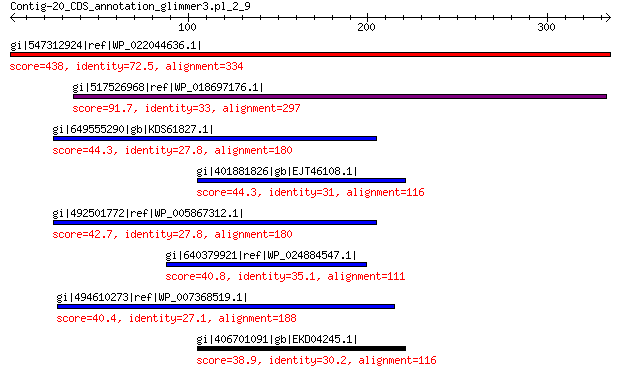
gi|547312924|ref|WP_022044636.1| putative uncharacterized protein 438 3e-150
gi|517526968|ref|WP_018697176.1| hypothetical protein 91.7 1e-17
gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 44.3 0.066
gi|401881826|gb|EJT46108.1| hypothetical protein A1Q1_05319 44.3 0.11
gi|492501772|ref|WP_005867312.1| hypothetical protein 42.7 0.19
gi|640379921|ref|WP_024884547.1| hypothetical protein 40.8 1.4
gi|494610273|ref|WP_007368519.1| hypothetical protein 40.4 1.5
gi|406701091|gb|EKD04245.1| hypothetical protein A1Q2_01464 38.9 5.2
>gi|547312924|ref|WP_022044636.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
gi|524208405|emb|CCZ76640.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
Length=328
Score = 438 bits (1126), Expect = 3e-150, Method: Compositional matrix adjust.
Identities = 242/337 (72%), Positives = 281/337 (83%), Gaps = 15/337 (4%)
Query 1 MSIlaglgaaaaSFAMKEGHNAIAQSRNEKNMALEHDYWKRRVNQLEEMNKPSRQVAKWR 60
MSILAGLGAAAASFAMKEGHNAIAQSRNEKNMALEHDYWKRRVNQLEEMNKPSRQVAKWR
Sbjct 1 MSILAGLGAAAASFAMKEGHNAIAQSRNEKNMALEHDYWKRRVNQLEEMNKPSRQVAKWR 60
Query 61 SAGIAPQAVFGNSPGGAGIATDASTPNSQTPMGSSDFNFVTTIAERQRMKNEKAIADATV 120
SAGIAPQAVFGNSPGGAGIATDAS+PNSQTPMGSSDFNFVTTIAERQRMKNEKAIADATV
Sbjct 61 SAGIAPQAVFGNSPGGAGIATDASSPNSQTPMGSSDFNFVTTIAERQRMKNEKAIADATV 120
Query 121 NKLNAEAEKLRGDTKDPNVTKDSQRLEFDWNLVKKQREQVQLAVDEINKEFQRAINEADV 180
+KLNAEA KLRGDTKDP VTKD Q+LEFDWN+VKKQRE+VQL VDEI+KEF+RA+NEAD+
Sbjct 121 DKLNAEAGKLRGDTKDPKVTKDLQQLEFDWNIVKKQRERVQLDVDEIDKEFRRAVNEADL 180
Query 181 QIKHGLYSETLAKIDKLIADKDVSEEMKQNLQKQRDLIESQISATQAQTDLIKaqtsatq 240
QIK G+YSETL+KIDKLIADK+VSEEMKQNLQKQRDLI++QI +T+AQT L K
Sbjct 181 QIKRGIYSETLSKIDKLIADKEVSEEMKQNLQKQRDLIDAQIDSTKAQTGLSK------- 233
Query 241 aqtETENALRDGRIKLTEREANKILADIGLSEARSLNE-YESLIKAMTGTQPASSLWGYI 299
AQT+TE+ LRDGR+KLT + +++L+ GL++ R E YE+ ++ + AS+ +
Sbjct 234 AQTKTEDTLRDGRVKLTGAQTSELLSMAGLNDVRRDREKYETFLR-LLDIDDASNGAEFA 292
Query 300 DRLIARGDSRLGGYENA--SDLREKLARALVRYIKAD 334
R+I +L G+ NA SD + K+ + I +D
Sbjct 293 QRII----RQLFGHMNADLSDYKRKMISDYLEKIWSD 325
>gi|517526968|ref|WP_018697176.1| hypothetical protein [Alistipes onderdonkii]
Length=364
Score = 91.7 bits (226), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 98/328 (30%), Positives = 149/328 (45%), Gaps = 58/328 (18%)
Query 36 HDYWKRRVNQLEEMNKPSRQVAKWRSAGIAPQAVFGNSPGGAGIATDA------------ 83
+YW + N PS Q AG++P F + G + D
Sbjct 64 QEYWNK-------YNSPSAQRIARMKAGMSP---FADESGVQAMGVDPGSYSGSSPSSQP 113
Query 84 -STP--NSQTPMGSSDFNFVTTIAERQRMKNEKAIADATVNKLNAEAEKLRGDTKDPNVT 140
+ P N+ +P+ + + V + ++ + + DA K AEA K + + T
Sbjct 114 FTQPGGNAMSPLSPAFASGVQQVLSARQAEANIQLTDANTAKTQAEAVKAQQENSLFAFT 173
Query 141 KDSQRLEFDWNLVKKQREQVQLAVDEINKEFQRAINEADV------------QIKHGLYS 188
K + E D L K Q + V E+ +F A +D+ Q K+ L S
Sbjct 174 KAA--AESD-ALSK----QFKATVAEVESQFAEAQALSDLAERNARIESIWAQAKNSLAS 226
Query 189 ETLAKIDKLIAD--KDVSEEMKQNLQKQRDLIESQISATQAQTDLIKaqtsatqaqtETE 246
+ D+L D KD + R+ ++SQ + QAQ + + AQ ETE
Sbjct 227 AAKSDADRLYLDFMKDAN----------RENVQSQTALNQAQAGTATSSAALMDAQRETE 276
Query 247 NALRDGRIKLTEREANKILADIGLSEARSLNEYESLIKAMTGTQPASSLWGYIDRLIARG 306
+ALR GRIKLTE +A LA GLSEAR+ EY LI+A+T T+ A+SLWG +DR + +
Sbjct 277 DALRSGRIKLTEEQARAALASAGLSEARAGREYNELIEALTNTRSANSLWGIVDRYVRKT 336
Query 307 DSRL-GGYENASD-LREKLARALVRYIK 332
++ L GG + +D LR L +A+ Y K
Sbjct 337 EAILPGGPQGKADELRFALIKAISNYSK 364
>gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649557306|gb|KDS63785.1| hypothetical protein M095_3404 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649559158|gb|KDS65545.1| hypothetical protein M096_4689 [Parabacteroides distasonis str.
3999B T(B) 6]
gi|649560567|gb|KDS66875.1| hypothetical protein M095_2448 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561016|gb|KDS67303.1| hypothetical protein M095_2410 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649562727|gb|KDS68911.1| hypothetical protein M096_3341 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=288
Score = 44.3 bits (103), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 50/212 (24%), Positives = 88/212 (42%), Gaps = 41/212 (19%)
Query 25 QSRNEKNMALEHDYWKRRVNQLEEMNKPSRQVAKWRSAGIAPQAVFGNSPGGAGIATDAS 84
Q NEK W N E N P++Q+A+ R+AG+ P V+GN G + S
Sbjct 47 QQENEKAYQRSLKMW----NLQNEYNSPTQQMARIRAAGLNPNLVYGNGVTG---NSAGS 99
Query 85 TP-------NSQTPMGSSDFNFVTTIAERQRMKNEKAIADATVNKLNAEAEKLRGD---- 133
TP N+ T +N + A Q + A V+ + A+ +R
Sbjct 100 TPQYEPAKFNAPTMQAYRGWNLGISDATSQYLAYR--TVKAQVDNMEAQNSLIRQQTATE 157
Query 134 -TKDPNVTKDSQRLEFDWNLVKKQRE-QVQLAVDEINK-------------------EFQ 172
T+ N+ + R EFD N+ K+ ++ V A+ E+N+ E
Sbjct 158 ATRQANIAASTSRSEFDLNMAKELKDVSVSSAIAEMNQKQAVAAQGWTKANREVVQYELD 217
Query 173 RAINEADVQIKHGLYSETLAKIDKLIADKDVS 204
+A+ + +++ + Y + L + +L D D++
Sbjct 218 KALFDNKIKLNNEKYLKALQSVRQLTQDNDIN 249
>gi|401881826|gb|EJT46108.1| hypothetical protein A1Q1_05319 [Trichosporon asahii var. asahii
CBS 2479]
Length=936
Score = 44.3 bits (103), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 36/117 (31%), Positives = 62/117 (53%), Gaps = 8/117 (7%)
Query 105 ERQRMKNEKAIADATVNKLNAEAEKLRGDTKDPNVTKDSQRLEFDWNLVKKQREQVQLAV 164
ER+R+K E A DA V +LNAE E+LR ++P+V + D + V + +EQ+ A+
Sbjct 632 ERRRLKKEVADRDAQVERLNAETERLREQPQEPSVRSEPG----DSSAVAELQEQLDAAM 687
Query 165 DEINKEFQRAINEADVQIKHGLYSETLAKIDKLIADK-DVSEEMKQNLQKQRDLIES 220
+++ I D++ +H + L + +AD D E+ ++L RD I+S
Sbjct 688 SKLSS---LEIQHQDLEAEHDDATAELEHTRQKLADTNDALEKASEDLYAARDAIDS 741
>gi|492501772|ref|WP_005867312.1| hypothetical protein [Parabacteroides distasonis]
gi|409230405|gb|EKN23269.1| hypothetical protein HMPREF1059_03254 [Parabacteroides distasonis
CL09T03C24]
Length=288
Score = 42.7 bits (99), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 50/212 (24%), Positives = 88/212 (42%), Gaps = 41/212 (19%)
Query 25 QSRNEKNMALEHDYWKRRVNQLEEMNKPSRQVAKWRSAGIAPQAVFGNSPGGAGIATDAS 84
Q NEK + W N E N P++Q+A+ R+AG+ P V+GN G + S
Sbjct 47 QQENEKAYQRSLNMW----NLQNEYNSPTQQMARIRAAGLNPNLVYGNGVTG---NSSGS 99
Query 85 TP-------NSQTPMGSSDFNFVTTIAERQRMKNEKAIADATVNKLNAEAEKLRGD---- 133
TP N+ T +N + A Q + A V+ + A+ +R
Sbjct 100 TPQYEPAKFNAPTMQAYRGWNLGISDAISQFLAYR--TVKAQVDNMEAQNSLIRQQTATE 157
Query 134 -TKDPNVTKDSQRLEFDWNLVKKQRE-QVQLAVDEINK-------------------EFQ 172
TK N+ + R EFD N+ K+ ++ V A+ ++N+ E
Sbjct 158 ATKQANIAASTSRSEFDLNMAKELKDVSVSSAIADMNQKQAGAAQGWTKANREVIQYELD 217
Query 173 RAINEADVQIKHGLYSETLAKIDKLIADKDVS 204
+A+ + +++ + Y L + +L D D++
Sbjct 218 KALFDNKIKLSNQEYLRVLQSVRQLQQDNDIN 249
>gi|640379921|ref|WP_024884547.1| hypothetical protein [Streptomyces sp. CNH189]
Length=1293
Score = 40.8 bits (94), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 39/126 (31%), Positives = 64/126 (51%), Gaps = 16/126 (13%)
Query 88 SQTPMGSSDFNFVTTIAERQRM------KNEKAIADAT--VNKLNAEAEKLRGDTKD--P 137
++ + +D T+AE +R+ K EK IADAT +L AEA + G +
Sbjct 931 TEDTITETDRLRTETVAEAERVRSESVAKAEKLIADATGDAERLRAEAAETVGSAQQHAE 990
Query 138 NVTKDSQRLEFDWN-----LVKKQREQVQLAVDEINKEFQRAINEADVQIKHGLYSETLA 192
+ +D++R++ D LV RE+ + +DE KE + +EA Q+ L +ET A
Sbjct 991 RIRRDAERVKTDAETEAERLVSGAREEAERTLDEARKEANKRRSEAAEQV-DTLITETTA 1049
Query 193 KIDKLI 198
+ DKL+
Sbjct 1050 EADKLL 1055
>gi|494610273|ref|WP_007368519.1| hypothetical protein [Prevotella multiformis]
gi|324988545|gb|EGC20508.1| hypothetical protein HMPREF9141_0987 [Prevotella multiformis
DSM 16608]
Length=437
Score = 40.4 bits (93), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 51/213 (24%), Positives = 90/213 (42%), Gaps = 27/213 (13%)
Query 27 RNEKNMALEHDYWKRRV-NQLEEMNKPSRQVAKWRSAGIAPQAVFGNSPGGAGIATDAST 85
+N+ M E + + R+ NQ+ + N P+ Q+ ++ AGI P GN G + S
Sbjct 109 QNQYQMFQEQNAFNERMWNQMNQYNSPAAQMQRYTDAGINPYIAAGNVQTGNAQSALQSA 168
Query 86 PNSQ---------TPMGSSDFNFVTTIAE--RQRMKNEKAIADAT--------VNKLN-A 125
P Q T MG + N I Q +N+ A+A A +++LN A
Sbjct 169 PAPQQHVAQVMPATGMGDAVQNSFAQIGNVISQFAQNQLALAQAKKTDAEASWIDRLNSA 228
Query 126 EAEKLRGDTKDPNVTKDSQRLEFDWNLVKKQREQVQLAVDEINKEFQRAINEADVQIKHG 185
+ KL +T N+ + L D+ + +L D ++ N D Q +
Sbjct 229 QMGKLGAETL--NIHNQNSLLGLDYQIKSDTLGNYKLLSDLSVQQAALTNNLVDAQTRKA 286
Query 186 LYSETLAKIDKLI----ADKDVSEEMKQNLQKQ 214
L+ LA ++ I +K V +E+ +++ +Q
Sbjct 287 LFESDLAMVESHIKAKYGEKQVLQEISESVSRQ 319
>gi|406701091|gb|EKD04245.1| hypothetical protein A1Q2_01464 [Trichosporon asahii var. asahii
CBS 8904]
Length=936
Score = 38.9 bits (89), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 35/118 (30%), Positives = 62/118 (53%), Gaps = 10/118 (8%)
Query 105 ERQRMKNEKAIADATVNKLNAEAEKLRGDTKDPNVTKDSQRLEFDWNLVKKQREQVQLAV 164
ER+R+K E A DA V +LNAE E+L ++P+V + D + V + +EQ+ A
Sbjct 632 ERRRLKKEVADRDAQVERLNAETERLHEQPQEPSVRSEPG----DSSAVAELQEQLDAAT 687
Query 165 DEINK-EFQRAINEADVQIKHGLYSETLAKIDKLIADKDVS-EEMKQNLQKQRDLIES 220
+++ E Q D++ +H + L + +AD + + E+ ++L RD I+S
Sbjct 688 SKLSSLELQ----HQDLEAEHDDATAELEHTRQKLADTNNALEKASEDLYAARDAIDS 741
Lambda K H a alpha
0.311 0.126 0.342 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1917593351550