bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-1_CDS_annotation_glimmer3.pl_2_1
Length=289
Score E
Sequences producing significant alignments: (Bits) Value
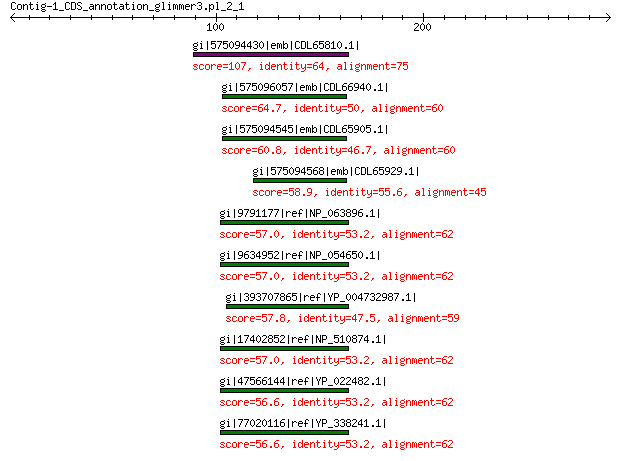
gi|575094430|emb|CDL65810.1| unnamed protein product 107 1e-23
gi|575096057|emb|CDL66940.1| unnamed protein product 64.7 8e-09
gi|575094545|emb|CDL65905.1| unnamed protein product 60.8 2e-07
gi|575094568|emb|CDL65929.1| unnamed protein product 58.9 1e-06
gi|9791177|ref|NP_063896.1| capsid protein VP2-related protein 57.0 1e-06
gi|9634952|ref|NP_054650.1| structural protein 57.0 1e-06
gi|393707865|ref|YP_004732987.1| structural protein VP2 57.8 1e-06
gi|17402852|ref|NP_510874.1| capsid protein VP2-related protein 57.0 1e-06
gi|47566144|ref|YP_022482.1| structural protein 56.6 1e-06
gi|77020116|ref|YP_338241.1| putative minor spike 56.6 1e-06
>gi|575094430|emb|CDL65810.1| unnamed protein product [uncultured bacterium]
Length=274
Score = 107 bits (266), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 48/75 (64%), Positives = 59/75 (79%), Gaps = 0/75 (0%)
Query 89 QGKYNSQSMLKQMGYNTLQAIMQGVYNHIENSVAMNYNSTEALANREWQEHMSSTAYQRA 148
QG YN +SML MGYNTL AI QG+YN I AM YNS +A+ +W+EHMS+T+YQRA
Sbjct 87 QGNYNFKSMLTSMGYNTLGAITQGIYNSISQGAAMQYNSAQAMRQMKWEEHMSNTSYQRA 146
Query 149 VEDMKKAGLNPILAF 163
+EDM+KAGLNPILA+
Sbjct 147 MEDMRKAGLNPILAY 161
>gi|575096057|emb|CDL66940.1| unnamed protein product [uncultured bacterium]
Length=275
Score = 64.7 bits (156), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 30/60 (50%), Positives = 43/60 (72%), Gaps = 0/60 (0%)
Query 103 YNTLQAIMQGVYNHIENSVAMNYNSTEALANREWQEHMSSTAYQRAVEDMKKAGLNPILA 162
+N QA +Q + +N+ AM +NS EA NR+WQE MS+TA+QR V+D+ AGLNP+L+
Sbjct 38 WNAEQAEIQRDWQEAQNAKAMQFNSMEAAKNRKWQEMMSNTAHQREVKDLMAAGLNPVLS 97
>gi|575094545|emb|CDL65905.1| unnamed protein product [uncultured bacterium]
Length=325
Score = 60.8 bits (146), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/60 (47%), Positives = 42/60 (70%), Gaps = 0/60 (0%)
Query 103 YNTLQAIMQGVYNHIENSVAMNYNSTEALANREWQEHMSSTAYQRAVEDMKKAGLNPILA 162
+N QA + +N++AM ++S EA NR+WQ +MS+TA+QR V D+K AGLNP+L+
Sbjct 37 FNASQAAANRNWQQQQNNIAMQFSSAEAAKNRDWQSYMSNTAHQREVADLKAAGLNPVLS 96
>gi|575094568|emb|CDL65929.1| unnamed protein product [uncultured bacterium]
Length=310
Score = 58.9 bits (141), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 25/45 (56%), Positives = 34/45 (76%), Gaps = 0/45 (0%)
Query 118 ENSVAMNYNSTEALANREWQEHMSSTAYQRAVEDMKKAGLNPILA 162
+N +AM +N+ EA NR WQE MS+TA+QR V D+ AGLNP+L+
Sbjct 71 QNRIAMQFNAAEAEKNRNWQEIMSNTAHQREVNDLMAAGLNPVLS 115
>gi|9791177|ref|NP_063896.1| capsid protein VP2-related protein [Chlamydia pneumoniae phage
CPAR39]
gi|7190962|gb|AAF39722.1| capsid protein VP2-related protein [Chlamydia pneumoniae phage
CPAR39]
Length=186
Score = 57.0 bits (136), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/65 (51%), Positives = 41/65 (63%), Gaps = 6/65 (9%)
Query 102 GYNTLQAIMQGVYNHIENSVAMNYNSTEALANRE---WQEHMSSTAYQRAVEDMKKAGLN 158
G + L I GV ++ A N+T RE +QE MS+TAYQRA+EDMKKAGLN
Sbjct 20 GLSFLPGIASGVLGYLG---AQKQNATAKQIAREQMAFQERMSNTAYQRAMEDMKKAGLN 76
Query 159 PILAF 163
P+LAF
Sbjct 77 PMLAF 81
>gi|9634952|ref|NP_054650.1| structural protein [Chlamydia phage 2]
gi|7406592|emb|CAB85592.1| structural protein [Chlamydia phage 2]
Length=186
Score = 57.0 bits (136), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/65 (51%), Positives = 41/65 (63%), Gaps = 6/65 (9%)
Query 102 GYNTLQAIMQGVYNHIENSVAMNYNSTEALANRE---WQEHMSSTAYQRAVEDMKKAGLN 158
G + L I GV ++ A N+T RE +QE MS+TAYQRA+EDMKKAGLN
Sbjct 20 GLSFLPGIASGVLGYLG---AQKQNATAKQIAREQMAFQERMSNTAYQRAMEDMKKAGLN 76
Query 159 PILAF 163
P+LAF
Sbjct 77 PMLAF 81
>gi|393707865|ref|YP_004732987.1| structural protein VP2 [Microviridae phi-CA82]
gi|311336637|gb|ADP89808.1| structural protein VP2 [Microviridae phi-CA82]
Length=234
Score = 57.8 bits (138), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/59 (47%), Positives = 38/59 (64%), Gaps = 0/59 (0%)
Query 105 TLQAIMQGVYNHIENSVAMNYNSTEALANREWQEHMSSTAYQRAVEDMKKAGLNPILAF 163
T QA Q +N + + +N+ EA NR+WQE MS+TA QR ++D +KAGLNPI A
Sbjct 9 TAQADKQNKWNAEQTEKSNQFNAQEAQKNRDWQEQMSNTALQRKMQDAEKAGLNPIFAL 67
>gi|17402852|ref|NP_510874.1| capsid protein VP2-related protein [Guinea pig Chlamydia phage]
Length=186
Score = 57.0 bits (136), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/65 (51%), Positives = 41/65 (63%), Gaps = 6/65 (9%)
Query 102 GYNTLQAIMQGVYNHIENSVAMNYNSTEALANRE---WQEHMSSTAYQRAVEDMKKAGLN 158
G + L I GV ++ A N+T RE +QE MS+TAYQRA+EDMKKAGLN
Sbjct 20 GLSFLPGIASGVLGYLG---AQKQNATAKQIAREQMAFQERMSNTAYQRAMEDMKKAGLN 76
Query 159 PILAF 163
P+LAF
Sbjct 77 PMLAF 81
>gi|47566144|ref|YP_022482.1| structural protein [Chlamydia phage 3]
gi|47522479|emb|CAD79480.1| structural protein [Chlamydia phage 3]
Length=186
Score = 56.6 bits (135), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/65 (51%), Positives = 41/65 (63%), Gaps = 6/65 (9%)
Query 102 GYNTLQAIMQGVYNHIENSVAMNYNSTEALANRE---WQEHMSSTAYQRAVEDMKKAGLN 158
G + L I GV ++ A N+T RE +QE MS+TAYQRA+EDMKKAGLN
Sbjct 20 GLSFLPGIASGVLGYLG---AQKQNATAKQIAREQMAFQERMSNTAYQRAMEDMKKAGLN 76
Query 159 PILAF 163
P+LAF
Sbjct 77 PMLAF 81
>gi|77020116|ref|YP_338241.1| putative minor spike [Chlamydia phage 4]
gi|59940015|gb|AAX12544.1| putative minor spike [Chlamydia phage 4]
Length=186
Score = 56.6 bits (135), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/65 (51%), Positives = 41/65 (63%), Gaps = 6/65 (9%)
Query 102 GYNTLQAIMQGVYNHIENSVAMNYNSTEALANRE---WQEHMSSTAYQRAVEDMKKAGLN 158
G + L I GV ++ A N+T RE +QE MS+TAYQRA+EDMKKAGLN
Sbjct 20 GLSFLPGIASGVLGYLG---AQKQNATAKQIAREQMAFQERMSNTAYQRAMEDMKKAGLN 76
Query 159 PILAF 163
P+LAF
Sbjct 77 PMLAF 81
Lambda K H a alpha
0.310 0.123 0.349 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1488295966443