bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-17_CDS_annotation_glimmer3.pl_2_8
Length=88
Score E
Sequences producing significant alignments: (Bits) Value
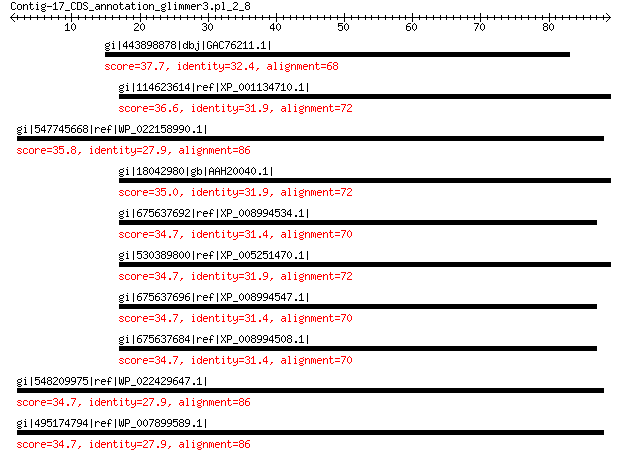
gi|443898878|dbj|GAC76211.1| hypothetical protein PANT_20d00003 37.7 0.12
gi|114623614|ref|XP_001134710.1| PREDICTED: KN motif and ankyrin... 36.6 1.3
gi|547745668|ref|WP_022158990.1| putative uncharacterized protein 35.8 2.2
gi|18042980|gb|AAH20040.1| KANK1 protein 35.0 4.6
gi|675637692|ref|XP_008994534.1| PREDICTED: KN motif and ankyrin... 34.7 5.8
gi|530389800|ref|XP_005251470.1| PREDICTED: KN motif and ankyrin... 34.7 5.9
gi|675637696|ref|XP_008994547.1| PREDICTED: KN motif and ankyrin... 34.7 6.1
gi|675637684|ref|XP_008994508.1| PREDICTED: KN motif and ankyrin... 34.7 6.2
gi|548209975|ref|WP_022429647.1| putative uncharacterized protein 34.7 6.5
gi|495174794|ref|WP_007899589.1| hypothetical protein 34.7 6.5
>gi|443898878|dbj|GAC76211.1| hypothetical protein PANT_20d00003 [Pseudozyma antarctica T-34]
Length=117
Score = 37.7 bits (86), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 22/76 (29%), Positives = 40/76 (53%), Gaps = 9/76 (12%)
Query 15 LPTTQEEKEYMIVIGKHL--------ATTEKFPTREAAEEAINSINWNLVTAMIYACKEA 66
+ TTQE+KE++ + K L AT+ P++ + + + S +W L+ A+I+AC A
Sbjct 1 MQTTQEQKEFLDNLTKVLGSTSAATCATSRSAPSKASGKATVPS-SWFLLLALIFACFSA 59
Query 67 DEYEKKLKRSAKKIDN 82
+ L + +DN
Sbjct 60 ASATRSLSMNQGPLDN 75
>gi|114623614|ref|XP_001134710.1| PREDICTED: KN motif and ankyrin repeat domain-containing protein
1 [Pan troglodytes]
Length=474
Score = 36.6 bits (83), Expect = 1.3, Method: Composition-based stats.
Identities = 23/72 (32%), Positives = 36/72 (50%), Gaps = 4/72 (6%)
Query 17 TTQEEKEYMIVIGKHLATTEKFPTREAAEEAINSINWNLVTAMIYACKEADEYEKKLKRS 76
T+Q E+E + GK +++ + FPT+E +N + + + A +YAC E LK
Sbjct 44 TSQSEQEVGTLEGKPISSLDAFPTQEGTLSPVNLTD-DQIAAGLYACTNN---ESTLKSI 99
Query 77 AKKIDNNTKKEG 88
KK D N G
Sbjct 100 MKKKDGNKDSNG 111
>gi|547745668|ref|WP_022158990.1| putative uncharacterized protein [Prevotella sp. CAG:520]
gi|524349976|emb|CDB05053.1| putative uncharacterized protein [Prevotella sp. CAG:520]
Length=748
Score = 35.8 bits (81), Expect = 2.2, Method: Composition-based stats.
Identities = 24/86 (28%), Positives = 45/86 (52%), Gaps = 16/86 (19%)
Query 2 EREFNDMKDVFRVLPTTQEEKEYMIVIGKHLATTEKFPTREAAEEAINSINWNLVTAMIY 61
E+E + +DVFR++ T++ + + ++ KH++ T+ F TR A +E N I
Sbjct 516 EKE-GEKEDVFRLMMRTRDPEIVLDLLMKHVSRTQSFMTRFAIDEFNNII---------- 564
Query 62 ACKEADEYEKKLKRSAKKIDNNTKKE 87
D E + R+ +K++N +KE
Sbjct 565 -----DGLENERTRTLRKVNNELRKE 585
>gi|18042980|gb|AAH20040.1| KANK1 protein [Homo sapiens]
Length=784
Score = 35.0 bits (79), Expect = 4.6, Method: Composition-based stats.
Identities = 23/72 (32%), Positives = 35/72 (49%), Gaps = 4/72 (6%)
Query 17 TTQEEKEYMIVIGKHLATTEKFPTREAAEEAINSINWNLVTAMIYACKEADEYEKKLKRS 76
T+Q E+E GK +++ + FPT+E +N + + + A +YAC E LK
Sbjct 354 TSQPEQEVGTSEGKPISSLDAFPTQEGTLSPVNLTD-DQIAAGLYACTNN---ESTLKSI 409
Query 77 AKKIDNNTKKEG 88
KK D N G
Sbjct 410 MKKKDGNKDSNG 421
>gi|675637692|ref|XP_008994534.1| PREDICTED: KN motif and ankyrin repeat domain-containing protein
1 isoform X5 [Callithrix jacchus]
Length=1312
Score = 34.7 bits (78), Expect = 5.8, Method: Composition-based stats.
Identities = 22/72 (31%), Positives = 37/72 (51%), Gaps = 3/72 (4%)
Query 17 TTQEEKEYMIVIGKHLATTEKFPTREAAEEAINSINWNLVTAMIYACKEADEYEKKL--K 74
T+Q E+E GK +++ + FPT+E +N + + + A +YAC + K + K
Sbjct 922 TSQPEQEVGTSEGKPISSLDAFPTQEGTLSPVNLTD-DQIAAGLYACTNNESTLKSIMKK 980
Query 75 RSAKKIDNNTKK 86
+ K N TKK
Sbjct 981 KDGNKDSNGTKK 992
>gi|530389800|ref|XP_005251470.1| PREDICTED: KN motif and ankyrin repeat domain-containing protein
1 isoform X4 [Homo sapiens]
Length=1284
Score = 34.7 bits (78), Expect = 5.9, Method: Composition-based stats.
Identities = 23/72 (32%), Positives = 35/72 (49%), Gaps = 4/72 (6%)
Query 17 TTQEEKEYMIVIGKHLATTEKFPTREAAEEAINSINWNLVTAMIYACKEADEYEKKLKRS 76
T+Q E+E GK +++ + FPT+E +N + + + A +YAC E LK
Sbjct 922 TSQPEQEVGTSEGKPISSLDAFPTQEGTLSPVNLTD-DQIAAGLYACTNN---ESTLKSI 977
Query 77 AKKIDNNTKKEG 88
KK D N G
Sbjct 978 MKKKDGNKDSNG 989
>gi|675637696|ref|XP_008994547.1| PREDICTED: KN motif and ankyrin repeat domain-containing protein
1 isoform X7 [Callithrix jacchus]
gi|675637698|ref|XP_008994555.1| PREDICTED: KN motif and ankyrin repeat domain-containing protein
1 isoform X7 [Callithrix jacchus]
gi|675637700|ref|XP_008994560.1| PREDICTED: KN motif and ankyrin repeat domain-containing protein
1 isoform X7 [Callithrix jacchus]
gi|675637702|ref|XP_008994566.1| PREDICTED: KN motif and ankyrin repeat domain-containing protein
1 isoform X7 [Callithrix jacchus]
gi|675637704|ref|XP_008994573.1| PREDICTED: KN motif and ankyrin repeat domain-containing protein
1 isoform X7 [Callithrix jacchus]
gi|675637708|ref|XP_008994583.1| PREDICTED: KN motif and ankyrin repeat domain-containing protein
1 isoform X7 [Callithrix jacchus]
Length=1235
Score = 34.7 bits (78), Expect = 6.1, Method: Composition-based stats.
Identities = 22/72 (31%), Positives = 37/72 (51%), Gaps = 3/72 (4%)
Query 17 TTQEEKEYMIVIGKHLATTEKFPTREAAEEAINSINWNLVTAMIYACKEADEYEKKL--K 74
T+Q E+E GK +++ + FPT+E +N + + + A +YAC + K + K
Sbjct 764 TSQPEQEVGTSEGKPISSLDAFPTQEGTLSPVNLTD-DQIAAGLYACTNNESTLKSIMKK 822
Query 75 RSAKKIDNNTKK 86
+ K N TKK
Sbjct 823 KDGNKDSNGTKK 834
>gi|675637684|ref|XP_008994508.1| PREDICTED: KN motif and ankyrin repeat domain-containing protein
1 isoform X1 [Callithrix jacchus]
Length=1393
Score = 34.7 bits (78), Expect = 6.2, Method: Composition-based stats.
Identities = 22/72 (31%), Positives = 37/72 (51%), Gaps = 3/72 (4%)
Query 17 TTQEEKEYMIVIGKHLATTEKFPTREAAEEAINSINWNLVTAMIYACKEADEYEKKL--K 74
T+Q E+E GK +++ + FPT+E +N + + + A +YAC + K + K
Sbjct 922 TSQPEQEVGTSEGKPISSLDAFPTQEGTLSPVNLTD-DQIAAGLYACTNNESTLKSIMKK 980
Query 75 RSAKKIDNNTKK 86
+ K N TKK
Sbjct 981 KDGNKDSNGTKK 992
>gi|548209975|ref|WP_022429647.1| putative uncharacterized protein [Prevotella stercorea CAG:629]
gi|524731096|emb|CDE34878.1| putative uncharacterized protein [Prevotella stercorea CAG:629]
Length=748
Score = 34.7 bits (78), Expect = 6.5, Method: Composition-based stats.
Identities = 24/86 (28%), Positives = 43/86 (50%), Gaps = 16/86 (19%)
Query 2 EREFNDMKDVFRVLPTTQEEKEYMIVIGKHLATTEKFPTREAAEEAINSINWNLVTAMIY 61
E+E + +DVFR++ T++ + + ++ KH++ T+ F TR A +E N I
Sbjct 516 EKE-GEKEDVFRLMMRTRDPEIVLDLLMKHVSRTQSFMTRFAIDEFNNII---------- 564
Query 62 ACKEADEYEKKLKRSAKKIDNNTKKE 87
D E + R+ +K+ N KE
Sbjct 565 -----DGLENERTRTLRKVSNELSKE 585
>gi|495174794|ref|WP_007899589.1| hypothetical protein [Prevotella stercorea]
gi|357558648|gb|EHJ40134.1| hypothetical protein HMPREF0673_01433 [Prevotella stercorea DSM
18206]
Length=748
Score = 34.7 bits (78), Expect = 6.5, Method: Composition-based stats.
Identities = 24/86 (28%), Positives = 43/86 (50%), Gaps = 16/86 (19%)
Query 2 EREFNDMKDVFRVLPTTQEEKEYMIVIGKHLATTEKFPTREAAEEAINSINWNLVTAMIY 61
E+E + +DVFR++ T++ + + ++ KH++ T+ F TR A +E N I
Sbjct 516 EKE-GEKEDVFRLMMRTRDPEIVLDLLMKHVSRTQSFMTRFAIDEFNNII---------- 564
Query 62 ACKEADEYEKKLKRSAKKIDNNTKKE 87
D E + R+ +K+ N KE
Sbjct 565 -----DGLENERTRTLRKVSNELSKE 585
Lambda K H a alpha
0.311 0.127 0.349 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 441619525350