bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-16_CDS_annotation_glimmer3.pl_2_1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
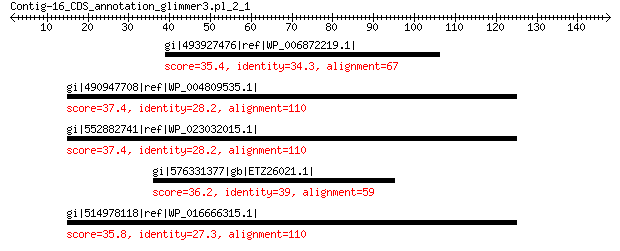
gi|493927476|ref|WP_006872219.1| hypothetical protein 35.4 2.0
gi|490947708|ref|WP_004809535.1| hypothetical protein 37.4 2.3
gi|552882741|ref|WP_023032015.1| MULTISPECIES: hypothetical protein 37.4 2.4
gi|576331377|gb|ETZ26021.1| transport family protein 36.2 6.0
gi|514978118|ref|WP_016666315.1| MULTISPECIES: hypothetical protein 35.8 7.8
>gi|493927476|ref|WP_006872219.1| hypothetical protein [Legionella drancourtii]
gi|363536305|gb|EHL29750.1| hypothetical protein LDG_8340 [Legionella drancourtii LLAP12]
Length=78
Score = 35.4 bits (80), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 23/75 (31%), Positives = 36/75 (48%), Gaps = 11/75 (15%)
Query 39 ETSDGDLIQC-----DMTQILLNQEKYRRLLGDMNIQNILAQ---MHPTQSTVMDGMTDE 90
E SD D++ D + LLNQE + RL MN+Q + Q M PT +++ + E
Sbjct 7 EISDSDMLNTVDELQDKKEYLLNQENFERL---MNVQKAVEQATEMRPTFKKLINSLVTE 63
Query 91 DRFACVISRHCQTMS 105
D + R + M+
Sbjct 64 DALTALTERLIKQMA 78
>gi|490947708|ref|WP_004809535.1| hypothetical protein [Propionibacterium avidum]
gi|348661485|gb|EGY78168.1| hypothetical protein HMPREF9153_0751 [Propionibacterium avidum
ATCC 25577]
Length=947
Score = 37.4 bits (85), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 31/114 (27%), Positives = 50/114 (44%), Gaps = 6/114 (5%)
Query 15 DYVPELVENHPCYQESVYDSVMYDETSDGDLIQCDMTQILLN----QEKYRRLLGDMNIQ 70
+ + E+VE + S + D + G L+Q D+ ++LL+ + K R LLG M
Sbjct 814 EAIDEMVETNLADIASSFSRSSRDLSGAGALLQADLNKVLLDLGDRKVKGRGLLGAMATS 873
Query 71 NILAQMHPTQSTVMDGMTDEDRFACVISRHCQTMSERQAVLQQLASEKSELTAY 124
+ Q + G E +A V R T+ RQA L+ + EL A+
Sbjct 874 SAKVGSADYQLAMASGAATE--YASVRGRDIDTLLLRQAQLKAATESQGELPAF 925
>gi|552882741|ref|WP_023032015.1| MULTISPECIES: hypothetical protein [Propionibacterium]
gi|550723104|gb|ERS22981.1| hypothetical protein HMPREF1301_00773 [Propionibacterium sp.
KPL2005]
gi|550729936|gb|ERS29662.1| hypothetical protein HMPREF1297_00481 [Propionibacterium sp.
KPL2000]
Length=947
Score = 37.4 bits (85), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 31/114 (27%), Positives = 50/114 (44%), Gaps = 6/114 (5%)
Query 15 DYVPELVENHPCYQESVYDSVMYDETSDGDLIQCDMTQILLN----QEKYRRLLGDMNIQ 70
+ + E+VE + S + D + G L+Q D+ ++LL+ + K R LLG M
Sbjct 814 EAIDEMVETNLADIASSFSRSSRDLSGAGALLQADLNKVLLDLGDRKVKGRGLLGAMATS 873
Query 71 NILAQMHPTQSTVMDGMTDEDRFACVISRHCQTMSERQAVLQQLASEKSELTAY 124
+ Q + G E +A V R T+ RQA L+ + EL A+
Sbjct 874 SAKVGSADYQLAMASGAATE--YASVRGRDIDTLLLRQAQLKAATESQGELPAF 925
>gi|576331377|gb|ETZ26021.1| transport family protein [Mycobacterium intracellulare MIN_052511_1280]
Length=959
Score = 36.2 bits (82), Expect = 6.0, Method: Composition-based stats.
Identities = 23/64 (36%), Positives = 34/64 (53%), Gaps = 5/64 (8%)
Query 36 MYDETSDGDLIQCDMTQILLNQEKYRRLLGDMN--IQNILAQMHPTQSTV---MDGMTDE 90
M D T+D + D+ ++ + E+ R LLG M+ ++A MH QSTV D + D
Sbjct 501 MKDRTADMLTMSDDLGAMVASMERMRGLLGQMSDTTHRMIADMHEMQSTVDQMRDHLADF 560
Query 91 DRFA 94
D FA
Sbjct 561 DDFA 564
>gi|514978118|ref|WP_016666315.1| MULTISPECIES: hypothetical protein [Propionibacterium]
gi|514204682|gb|EPH00491.1| hypothetical protein HMPREF1485_00816 [Propionibacterium sp.
HGH0353]
gi|537782525|gb|ERF58339.1| hypothetical protein H639_04232 [Propionibacterium avidum TM16]
gi|550741599|gb|ERS41152.1| hypothetical protein HMPREF1271_00782 [Propionibacterium sp.
KPL1838]
gi|550769982|gb|ERS68141.1| hypothetical protein HMPREF1279_00500 [Propionibacterium sp.
KPL1852]
Length=913
Score = 35.8 bits (81), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 50/114 (44%), Gaps = 6/114 (5%)
Query 15 DYVPELVENHPCYQESVYDSVMYDETSDGDLIQCDMTQILLN----QEKYRRLLGDMNIQ 70
+ + E+V+ + S + D + G L+Q D+ ++LL+ + K R LLG M
Sbjct 780 EAIDEMVKTNLADIASSFSRSSRDLSGAGALLQADLNKVLLDLGDRKVKGRGLLGAMATS 839
Query 71 NILAQMHPTQSTVMDGMTDEDRFACVISRHCQTMSERQAVLQQLASEKSELTAY 124
+ Q + G E +A V R T+ RQA L+ + EL A+
Sbjct 840 SAKVGSADYQLAMASGAATE--YASVRGRDIDTLLLRQAQLKAATESQGELPAF 891
Lambda K H a alpha
0.317 0.129 0.368 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 436430036016