bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-15_CDS_annotation_glimmer3.pl_2_5
Length=589
Score E
Sequences producing significant alignments: (Bits) Value
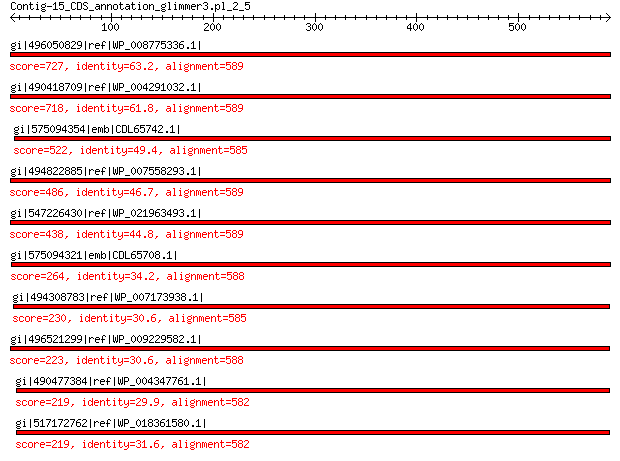
gi|496050829|ref|WP_008775336.1| hypothetical protein 727 0.0
gi|490418709|ref|WP_004291032.1| hypothetical protein 718 0.0
gi|575094354|emb|CDL65742.1| unnamed protein product 522 3e-175
gi|494822885|ref|WP_007558293.1| hypothetical protein 486 4e-161
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 438 5e-143
gi|575094321|emb|CDL65708.1| unnamed protein product 264 2e-75
gi|494308783|ref|WP_007173938.1| hypothetical protein 230 6e-64
gi|496521299|ref|WP_009229582.1| capsid protein 223 2e-61
gi|490477384|ref|WP_004347761.1| capsid protein 219 1e-59
gi|517172762|ref|WP_018361580.1| hypothetical protein 219 1e-59
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 727 bits (1876), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 372/609 (61%), Positives = 447/609 (73%), Gaps = 49/609 (8%)
Query 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEVLPGDTFKIDLKSFTRTQP 60
MANIMSLKSLRNKTSRNGFDLSSKRNFTAK GELLPVK WEVLPGD + IDLKSFTRTQP
Sbjct 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKPGELLPVKCWEVLPGDKWSIDLKSFTRTQP 60
Query 61 LNTAAFARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHALDSSPLNVVKLDGSMPFTD 120
LNTAAFARMREYYDFYFVPY+LLWNKANT LTQMYDNPQHA P L G MP
Sbjct 61 LNTAAFARMREYYDFYFVPYNLLWNKANTVLTQMYDNPQHATSYIPSANQALAGVMPNVT 120
Query 121 LSSISRYLNSLASNSTAVTN-KANYFGYNRALCSAKLMECLGYGNLYYYAESTSNTFAKR 179
I+ YLN +A + T + + NYFGY+R+L +AKL+E LGYGN Y YA S +NT+ K
Sbjct 121 CKGIADYLNLVAPDVTTTNSYEKNYFGYSRSLGTAKLLEYLGYGNFYTYATSKNNTWTKS 180
Query 180 PLHYNLNVSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYMPYSTGSS----GMMLSFN 235
PL NL ++++ +LAYQKIYAD+ RDSQWE+VSPSCFNVDY+ + S+ M+
Sbjct 181 PLSSNLQLNIYGVLAYQKIYADHIRDSQWEKVSPSCFNVDYLSGTVDSAMTIDSMITGQG 240
Query 236 YSDFYENYSMFDLRYCNWQKDLFHGVVPNQQYGDVASISMSVPVVAGSSAAlinsritss 295
++ FY +MFDLRYCNWQKDLFHGV+P QQYGD A++++++ V + +
Sbjct 241 FAPFY---NMFDLRYCNWQKDLFHGVLPRQQYGDTAAVNVNLSNVLSAQYMVQT------ 291
Query 296 nstttlrFPTDPAIPDATPLLTHP---------------SFSILALRQAEFLQKWKEITQ 340
PD P+ P +F++LALRQAEFLQKWKEITQ
Sbjct 292 --------------PDGDPVGGSPFSSTGVNLQTVNGSGTFTVLALRQAEFLQKWKEITQ 337
Query 341 SGNKDYKEQVEKHWNVSPGDGFSEMCTYLGGISSSLDINEVVNQNITGSNAADIAGKGTG 400
SGNKDYK+Q+EKHWNVS G+ +SEM YLGG ++SLDINEVVN NITGSNAADIAGKG
Sbjct 338 SGNKDYKDQIEKHWNVSVGEAYSEMSLYLGGTTASLDINEVVNNNITGSNAADIAGKGVV 397
Query 401 VSNGVINFNSQGRYGVVMCIYHCLPLIDYTTDFVSPSVTRVNAADFAIPEFDRVGMQTVP 460
V NG I+F++ RYG++MCIYH LPL+DYTTD V+P+ T++N+ DFAIPEFDRVGM++VP
Sbjct 398 VGNGRISFDAGERYGLIMCIYHSLPLLDYTTDLVNPAFTKINSTDFAIPEFDRVGMESVP 457
Query 461 LSYVSNGPLSVLPLSIPNEIGYAPRYIDYKTDIDTSIGAFKTSLKNWVISYDNQSLANQF 520
L + N PL + +GYAPRYI YKTD+D+S+GAFKT+LK+WV+SYDNQS+ NQ
Sbjct 458 LVSLMN-PLQSSYNVGSSILGYAPRYISYKTDVDSSVGAFKTTLKSWVMSYDNQSVINQL 516
Query 521 GYSVETPESPVPNPANSSWNYTLFKVNPNSLNPLFAVEADSSIDTDQFLCSTFFDVKVVR 580
Y + SP + NYT FKVNPN ++PLFAV A +SIDTDQFLCS+FFDVKVVR
Sbjct 517 NYQDDPNNSP-----GTLVNYTNFKVNPNCVDPLFAVAASNSIDTDQFLCSSFFDVKVVR 571
Query 581 NLDTDGLPY 589
NLDTDGLPY
Sbjct 572 NLDTDGLPY 580
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 718 bits (1854), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 364/598 (61%), Positives = 436/598 (73%), Gaps = 29/598 (5%)
Query 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEVLPGDTFKIDLKSFTRTQP 60
MANIMSLKS+RNK SRNGFDLS K+NFTAKAGELLPV EVLPGDTFKI+LK+FTRTQP
Sbjct 1 MANIMSLKSIRNKPSRNGFDLSFKKNFTAKAGELLPVMVKEVLPGDTFKINLKAFTRTQP 60
Query 61 LNTAAFARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHALDSSPLNVVKLDGSMPFTD 120
+NTAAFAR+REYYDF+FVPYDLLWNKANT LTQMYDNPQHA+ P L G MP+
Sbjct 61 VNTAAFARIREYYDFFFVPYDLLWNKANTVLTQMYDNPQHAVSIDPTRNFVLSGEMPYMT 120
Query 121 LSSISRYLNSLASNSTAVTNKANYFGYNRALCSAKLMECLGYGNLYYYAESTSNTFAKRP 180
+I+ Y+N+L++ S K+NYFGYNR+ S KL+E LGYGN Y ++ + P
Sbjct 121 SEAIASYINALSTASALADYKSNYFGYNRSKSSVKLLEYLGYGN---YESFLTDDWNTAP 177
Query 181 LHYNLNVSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYMPYSTGSSGMMLSFNYS-DF 239
L NLN ++F LLAYQKIY+D+YRDSQWERVSPS FNVDY+ S M L YS +F
Sbjct 178 LMANLNHNIFGLLAYQKIYSDFYRDSQWERVSPSTFNVDYL----DGSSMNLDNAYSTEF 233
Query 240 YENYSMFDLRYCNWQKDLFHGVVPNQQYGDVASISMSVPVVAGSSAAlinsritssnstt 299
Y+NY+ FDLRYCNWQKDLFHGV+P+QQYG+ A S++ P V G +
Sbjct 234 YQNYNFFDLRYCNWQKDLFHGVLPHQQYGETAVASIT-PDVTGK-----------LTLSN 281
Query 300 tlrFPTDPAIPDATPLLTHPSF------SILALRQAEFLQKWKEITQSGNKDYKEQVEKH 353
T P T P+F SIL LRQAEFLQKWKEITQSGNKDYK+Q+EKH
Sbjct 282 FSTVGTSPTTASGTATKNLPAFDTVGDLSILVLRQAEFLQKWKEITQSGNKDYKDQLEKH 341
Query 354 WNVSPGDGFSEMCTYLGGISSSLDINEVVNQNITGSNAADIAGKGTGVSNGVINFNSQGR 413
W VS GDGFSE+CTYLGG+SSS+DINEV+N NITGS AADIAGKG GV+NG INFNS GR
Sbjct 342 WGVSVGDGFSELCTYLGGVSSSIDINEVINTNITGSAAADIAGKGVGVANGEINFNSNGR 401
Query 414 YGVVMCIYHCLPLIDYTTDFVSPSVTRVNAADFAIPEFDRVGMQTVPLSYVSNGPLSVLP 473
YG++MCIYHCLPL+DYTTD + P+ +VN+ D+AIPEFDRVGMQ++PL + N PL
Sbjct 402 YGLIMCIYHCLPLLDYTTDMLDPAFLKVNSTDYAIPEFDRVGMQSMPLVQLMN-PLRSFA 460
Query 474 LSIPNEIGYAPRYIDYKTDIDTSIGAFKTSLKNWVISYDNQSLANQFGYSVETP--ESPV 531
+ +GY PRYIDYKT +D S+G FK +L +WVISY N S+ Q + P E
Sbjct 461 NASGLVLGYVPRYIDYKTSVDQSVGGFKRTLNSWVISYGNISVLKQVTLPNDAPPIEPSE 520
Query 532 PNPANSSWNYTLFKVNPNSLNPLFAVEADSSIDTDQFLCSTFFDVKVVRNLDTDGLPY 589
P P+ + N+T FKVNP+ L+P+FAV+A +TDQFLCS+FFD+K VRNLDTDGLPY
Sbjct 521 PVPSVAPMNFTFFKVNPDCLDPIFAVQAGDDTNTDQFLCSSFFDIKAVRNLDTDGLPY 578
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 522 bits (1345), Expect = 3e-175, Method: Compositional matrix adjust.
Identities = 289/631 (46%), Positives = 390/631 (62%), Gaps = 62/631 (10%)
Query 5 MSLKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEVLPGDTFKIDLKSFTRTQPLNTA 64
MS+ ++N+ SRNGFDLS K+NFTAKAGELLPV T VLPGD+F I+L+SFTRTQPLNT+
Sbjct 1 MSMADIKNRPSRNGFDLSFKKNFTAKAGELLPVMTKVVLPGDSFNINLRSFTRTQPLNTS 60
Query 65 AFARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHALDSSPLNVVKLDGSMPFTDLSSI 124
AFARMREYYDFYFVP++ +WNK ++ +TQM N QHA + + L G MP+ I
Sbjct 61 AFARMREYYDFYFVPFEQMWNKFDSCITQMNANVQHASGPTLDDNTPLSGRMPYFTSEQI 120
Query 125 SRYLNSLASNSTAVTNKANYFGYNRALCSAKLMECLGYGNLYYYAESTSNTFAKRPLHYN 184
+ YLN A+ + + N FG+NR+ + KL++ LGYG+ Y +S +NT++ +PL YN
Sbjct 121 ADYLNDQATAA-----RKNPFGFNRSTLTCKLLQYLGYGD-YNSFDSETNTWSAKPLLYN 174
Query 185 LNVSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYMPYSTGSSGMMLSFNYSDFYENYS 244
L +S F LLAYQKIY+D+YR +QWE+ +PS FN+DY+ G+S + + +N +
Sbjct 175 LELSPFPLLAYQKIYSDFYRYTQWEKTNPSTFNLDYI---KGTSDLQMDLTGLPSDDN-N 230
Query 245 MFDLRYCNWQKDLFHG--------------------VVPNQQYGDVASISMSVPVVAGSS 284
FD+RYCN+QKD+FHG V+ N G + S P G+S
Sbjct 231 FFDIRYCNYQKDMFHGVLPVAQYGSASVVPINGQLNVISNGDSGPIFKTSTPDPGTPGTS 290
Query 285 ----------------AAlinsritssnstttlrFPTDPAIPDATPLLTHPSF------- 321
+ + S + FP++ + + L +P+
Sbjct 291 YVTVGGNIGVDNRSFGVSGSTLNVGKSADPSGYGFPSNAST--RSLLWENPNLIIENNQG 348
Query 322 ---SILALRQAEFLQKWKEITQSGNKDYKEQVEKHWNVSPGDGFSEMCTYLGGISSSLDI 378
ILALRQAEFLQKWKE++ SG +DYK Q+EKHW + D S YLGG ++SLDI
Sbjct 349 FYVPILALRQAEFLQKWKEVSVSGEEDYKSQIEKHWGIKVSDFLSHQARYLGGCATSLDI 408
Query 379 NEVVNQNITGSNAADIAGKGTGVSNGVINFNSQGRYGVVMCIYHCLPLIDYTTDFVSPSV 438
NEV+N NITG NAADIAGKGT NG I F S+G YG++MCIYH LP++DY V S
Sbjct 409 NEVINNNITGDNAADIAGKGTFTGNGSIRFESKGEYGIIMCIYHVLPIVDYVGSGVDHSC 468
Query 439 TRVNAADFAIPEFDRVGMQTVPLSYVSNGPLSVLPLSIPNEIGYAPRYIDYKTDIDTSIG 498
T V+A F IPE D++GM++VPL N S +GYAPRYID+KT +D S+G
Sbjct 469 TLVDATSFPIPELDQIGMESVPLVRAMNPVKESDTPSADTFLGYAPRYIDWKTSVDRSVG 528
Query 499 AFKTSLKNWVISYDNQSLANQFGYSVETPESPVPNPANSSWNYTLFKVNPNSLNPLFAVE 558
F SL+ W + ++ L + S+ P +P P + + + FKVNP+ ++PLFAV
Sbjct 529 DFADSLRTWCLPVGDKELTS--ANSLNFPSNPNVEPDSIAAGF--FKVNPSIVDPLFAVV 584
Query 559 ADSSIDTDQFLCSTFFDVKVVRNLDTDGLPY 589
ADS++ TD+FLCS+FFDVKVVRNLD +GLPY
Sbjct 585 ADSTVKTDEFLCSSFFDVKVVRNLDVNGLPY 615
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 486 bits (1251), Expect = 4e-161, Method: Compositional matrix adjust.
Identities = 275/622 (44%), Positives = 371/622 (60%), Gaps = 49/622 (8%)
Query 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEVLPGDTFKIDLKSFTRTQP 60
MANIMS+KS+RNK +R G+DL+ K NFTAKAG L+PV VLP D +KSF RTQP
Sbjct 8 MANIMSMKSVRNKPTRAGYDLTQKINFTAKAGSLIPVWWTPVLPFDDLNATVKSFVRTQP 67
Query 61 LNTAAFARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHALDSSPLNVVKLDGSMPFTD 120
LNTAAFARMR Y+DFYFVP+ +WNK T++TQM N HA + V L +P+
Sbjct 68 LNTAAFARMRGYFDFYFVPFRQMWNKFPTAITQMRTNLLHASGPVLADNVPLSDELPYFT 127
Query 121 LSSISRYLNSLASNSTAVTNKANYFGYNRALCSAKLMECLGYGNLYYY----AESTSNTF 176
++ Y+ SLA + N FGY RA ++E LGYG+ Y Y A T+
Sbjct 128 AEQVADYIVSLA-------DSKNQFGYYRAWLVCIILEYLGYGDFYPYIVEAAGGEGATW 180
Query 177 AKRPLHYNLNVSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYMPYSTGSSGMMLSFNY 236
A RP+ NL S F L AYQKIYAD+ R +QWER +PS FN+DY+ S + + L F
Sbjct 181 ATRPMLNNLKFSPFPLFAYQKIYADFNRYTQWERSNPSTFNIDYI--SGSADSLQLDFTV 238
Query 237 SDFYENYSMFDLRYCNWQKDLFHGVVPNQQYGDVASI--SMSVPVVAGSSA--------A 286
F +++++FD+RY NWQ+DL HG +P QYG+ +++ S S+ VV G +
Sbjct 239 EGFKDSFNLFDMRYSNWQRDLLHGTIPQAQYGEASAVPVSGSMQVVEGPTPPAFTTGQDG 298
Query 287 linsritssnstttlrFPTDPAIPDATPLLTHPS-------------FSILALRQAEFLQ 333
+ + ++ ++ ++ L + + SILALR+AE Q
Sbjct 299 VAFLNGNVTIQGSSGYLQAQTSVGESRILRFNNTNSGLIVEGDSSFGVSILALRRAEAAQ 358
Query 334 KWKEITQSGNKDYKEQVEKHWNVSPGDGFSEMCTYLGGISSSLDINEVVNQNITGSNAAD 393
KWKE+ + +DY Q+E HW S +S+MC +LG I+ L INEVVN NITG NAAD
Sbjct 359 KWKEVALASEEDYPSQIEAHWGQSVNKAYSDMCQWLGSINIDLSINEVVNNNITGENAAD 418
Query 394 IAGKGTGVSNGVINFNSQGRYGVVMCIYHCLPLIDYTTDFVSPSVTRVNAADFAIPEFDR 453
IAGKGT NG INFN G+YG+VMC++H LP +DY T T N DF IPEFD+
Sbjct 419 IAGKGTMSGNGSINFNVGGQYGIVMCVFHVLPQLDYITSAPHFGTTLTNVLDFPIPEFDK 478
Query 454 VGMQTVPLSYVSN------GPLSVLPLSIPNEIGYAPRYIDYKTDIDTSIGAFKTSLKNW 507
+GM+ VP+ N G V P GYAP+Y ++KT +D S+G F+ SLK W
Sbjct 479 IGMEQVPVIRGLNPVKPKDGDFKVSPNLY---FGYAPQYYNWKTTLDKSMGEFRRSLKTW 535
Query 508 VISYDNQSLANQFGYSVETPESPVPNPANSSWNYTLFKVNPNSLNPLFAVEADSSIDTDQ 567
+I +D+++L SV+ P++ PN S FKV+P+ L+ LFAV+A+S ++TDQ
Sbjct 536 IIPFDDEALLA--ADSVDFPDN--PNVEADSVKAGFFKVSPSVLDNLFAVKANSDLNTDQ 591
Query 568 FLCSTFFDVKVVRNLDTDGLPY 589
FLCST FDV VVR+LD +GLPY
Sbjct 592 FLCSTLFDVNVVRSLDPNGLPY 613
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 438 bits (1127), Expect = 5e-143, Method: Compositional matrix adjust.
Identities = 264/596 (44%), Positives = 362/596 (61%), Gaps = 30/596 (5%)
Query 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEVLPGDTFKIDLKSFTRTQP 60
M+++MSL +L+N RNGFDLS K FTAK GELLP+ EV PGD F I ++FTRTQP
Sbjct 1 MSSVMSLTALKNSVKRNGFDLSFKNAFTAKVGELLPIMCKEVYPGDKFNIRGQAFTRTQP 60
Query 61 LNTAAFARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHALDSSPLNVVKLDGSMPFTD 120
+N+AA++R+REYYDFYFVPY LLWN A T T M D P HA D ++ V L P+
Sbjct 61 VNSAAYSRLREYYDFYFVPYRLLWNMAPTFFTNMPD-PHHAADL--VSSVNLSQRHPWFT 117
Query 121 LSSISRYLNSLASNSTAVTN-KANYFGYNRALCSAKLMECL--GYGNLYYYAESTSNTFA 177
I YL +L S S A + N+FG++R S KL+ L G+G Y + S++
Sbjct 118 FFDIMEYLGNLNSLSGAYEKYQKNFFGFSRVELSVKLLNYLNYGFGKDYESVKVPSDS-- 175
Query 178 KRPLHYNLNVSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYM-PYSTGSSGMMLSFNY 236
++ +S F LLAYQKI DY+RD QW+ +P +N+DY+ S+G M SF
Sbjct 176 -----DDIVLSPFPLLAYQKICEDYFRDDQWQSAAPYRYNLDYLYGKSSGFHIPMSSFT- 229
Query 237 SDFYENYSMFDLRYCNWQKDLFHGVVPNQQYGDVASISMSVPVVAGSSAAlinsritssn 296
+D ++N +MFDL YCN+QKD F G++P QYGDV S++ P+ +S +S
Sbjct 230 NDAFKNPTMFDLNYCNFQKDYFTGMLPRAQYGDV---SVASPIFGDLDIGDSSSLTFASA 286
Query 297 stttlrFPTDPAIPDATPLLTHPSFSILALRQAEFLQKWKEITQSGNKDYKEQVEKHWNV 356
+ T S+LALRQAE LQKW+EI QSG DY+ Q++KH+NV
Sbjct 287 PQQGANTIQSGVLVVNNNSNTTAGLSVLALRQAECLQKWREIAQSGKMDYQTQMQKHFNV 346
Query 357 SPGDGFSEMCTYLGGISSSLDINEVVNQNITGSNAADIAGKGTGVSNG-VINFNSQGRYG 415
SP S C YLGG +S+LDI+EVVN N+TG N ADI GKGTG NG ++F S +G
Sbjct 347 SPSATLSGHCKYLGGWTSNLDISEVVNTNLTGDNQADIQGKGTGTLNGNKVDFESS-EHG 405
Query 416 VVMCIYHCLPLIDYTTDFVSPSVTRVNAADFAIPEFDRVGMQTVPLSYVSNGPLSVLPLS 475
++MCIYHCLPL+D++ + ++ + D+AIPEFD VGMQ + S + G L LP S
Sbjct 406 IIMCIYHCLPLLDWSINRIARQNFKTTFTDYAIPEFDSVGMQQLYPSEMIFG-LEDLP-S 463
Query 476 IPNEI--GYAPRYIDYKTDIDTSIGAFKTSLKNWVISYDNQSLANQFGYSVETPESPVPN 533
P+ I GY PRY D KT ID G+F +L +WV + ++ Y ++
Sbjct 464 DPSSINMGYVPRYADLKTSIDEIHGSFIDTLVSWVSPLTDSYIS---AYRQACKDAGF-- 518
Query 534 PANSSWNYTLFKVNPNSLNPLFAVEADSSIDTDQFLCSTFFDVKVVRNLDTDGLPY 589
++ + Y FKVNP+ ++ +F V+ADS+I+TDQ L +++FD+K VRN D +GLPY
Sbjct 519 -SDITMTYNFFKVNPHIVDNIFGVKADSTINTDQLLINSYFDIKAVRNFDYNGLPY 573
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 264 bits (674), Expect = 2e-75, Method: Compositional matrix adjust.
Identities = 201/656 (31%), Positives = 306/656 (47%), Gaps = 88/656 (13%)
Query 2 ANIMSLKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEVLPGDTFKIDLKSFTRTQPL 61
+NIM L L+NK SRN FDLS + FTAK GELLP E+ PGD+ K+ FTRT PL
Sbjct 5 SNIMGLHGLKNKPSRNSFDLSHRNMFTAKVGELLPCFVQELNPGDSVKVSSSYFTRTAPL 64
Query 62 NTAAFARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHA----LDSSPLNVVKLDGSMP 117
+ AF R+RE ++FVPY LW ++ + M N + SS + K+ MP
Sbjct 65 QSNAFTRLRENVQYFFVPYSALWKYFDSQVLNMTKNANGGDISRIASSLVGNQKVTTQMP 124
Query 118 FTDLSSISRYLNSLASNSTAVTNKANYFGYNRALC-----SAKLMECLGYGNLYYYAEST 172
+ ++ YL + ST ++ + +NR C SAKL++ LGYGN + E
Sbjct 125 CVNYKTLHAYLLKFINRSTVGSDGSVGPEFNRG-CYRHAESAKLLQLLGYGN---FPEQF 180
Query 173 SNTFAKRPLH-----------YNLN--VSLFNLLAYQKIYADYYRDSQWERVSPSCFNVD 219
+N H YN + +S+F LLAY KI D+Y QW+ + S NVD
Sbjct 181 ANFKVNNDKHNQSGQNFKDVTYNNSPYLSIFRLLAYHKICNDHYLYRQWQPYNASLCNVD 240
Query 220 YMPYSTGSSGMMLSFNYSDF--------YENYSMFDLRYCNWQKDLFHGVVPNQQYGDVA 271
Y+ T +S +LS + + E ++ D+R+ N D F GV+P Q+G +
Sbjct 241 YL---TPNSSSLLSIDDALLSIPDDSIKAEKLNLLDMRFSNLPLDYFTGVLPTSQFGSES 297
Query 272 SISMSVPVVAGSS-------------------------AAlinsritssnstttlrFPTD 306
+++++ +GS+ A + +++ D
Sbjct 298 VVNLNLGNASGSAVLNGTTSKDSGRWRTTTGEWEMEQRVASSANGNLKLDNSNGTFISHD 357
Query 307 PAIPDATPLLTHPS--FSILALRQAEFLQKWKEITQSGNKDYKEQVEKHWNVSPGDGFSE 364
+ T S SI+ALR A QK+KEI + + D++ QVE H+ + P D +E
Sbjct 358 HTFSGNVAINTSLSGNLSIIALRNALAAQKYKEIQLANDVDFQSQVEAHFGIKP-DEKNE 416
Query 365 MCTYLGGISSSLDINEVVNQNITGSNAADIAGKGTGVSNGVINFNSQGRYGVVMCIYHCL 424
++GG SS ++INE +NQN++G N A G + I F ++ YGVV+ IY C
Sbjct 417 NSLFIGGSSSMININEQINQNLSGDNKATYGAAPQGNGSASIKFTAK-TYGVVIGIYRCT 475
Query 425 PLIDYTTDFVSPSVTRVNAADFAIPEFDRVGMQTVPLSYVS-----NGPLSVLPLS---- 475
P++D+ + ++ + +A+DF IPE D +GMQ V+ N +
Sbjct 476 PVLDFAHLGIDRTLFKTDASDFVIPEMDSIGMQQTFRCEVAAPAPYNDEFKAFRVGDGSS 535
Query 476 --IPNEIGYAPRYIDYKTDIDTSIGAFKTSLKNWVISYDNQSLANQFGYSVETPESPVPN 533
+ GYAPRY ++KT D GAF SLK+WV + ++ N + +P
Sbjct 536 PDMSETYGYAPRYSEFKTSYDRYNGAFCHSLKSWVTGINFDAIQNNVWNTWAGINAP--- 592
Query 534 PANSSWNYTLFKVNPNSLNPLFAVEADSSIDTDQFLCSTFFDVKVVRNLDTDGLPY 589
+F P+ + LF V + ++ D DQ RNL GLPY
Sbjct 593 --------NMFACRPDIVKNLFLVSSTNNSDDDQLYVGMVNMCYATRNLSRYGLPY 640
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 230 bits (587), Expect = 6e-64, Method: Compositional matrix adjust.
Identities = 179/605 (30%), Positives = 289/605 (48%), Gaps = 78/605 (13%)
Query 4 IMSLKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEVLPGDTFKIDLKSFTRTQPLNT 63
I +K+ R +RN FDLS + FTA AG LLPV +++P D +I+ + F RT P+NT
Sbjct 5 IPKIKATRPNRNRNAFDLSQRHLFTAHAGMLLPVLNLDLIPHDHVEINAQDFMRTLPMNT 64
Query 64 AAFARMREYYDFYFVPYDLLWNKANTSLTQMYD-----NPQHALDSSPLNVVKLDGSMPF 118
AAFA MR Y+F+FVPY LW + + +T M D N +SPL V P+
Sbjct 65 AAFASMRGVYEFFFVPYHQLWAQFDQFITGMNDFHSSANKSIQGGTSPLQV-------PY 117
Query 119 TDLSSISRYLNSLASNSTAVTNKANY-FGYNRALCSAKLMECLGYGNLY-YYAESTSNTF 176
++ S+ LN+ + + T+ Y F Y + +L++ LGYG + + + +
Sbjct 118 FNVDSVFNSLNTGKESGSGSTDDLQYKFKYG----AFRLLDLLGYGRKFDSFGTAYPDNV 173
Query 177 AKRPLHYNLNVSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYMPYSTGSSGMMLSFNY 236
+ + + N S+F +LAY KIY DYYR+S +E FN D G++ +
Sbjct 174 SGLKNNLDYNCSVFRILAYNKIYQDYYRNSNYENFDTDSFNFDKF-----KGGLVDAKVV 228
Query 237 SDFYENYSMFDLRYCNWQKDLFHGVVPNQQYGDVASISMSVPVVAGSSAAlinsritssn 296
+D +F LRY N Q D F + +Q + S + + V + A + + +
Sbjct 229 AD------LFKLRYRNAQTDYFTNLRQSQLF----SFTTAFEDVDNINIAPRDYVKSDGS 278
Query 297 stttlrFPTDPAIPDATPLLTHPSFSILALRQAEFLQKWKEITQSGNKDYKEQVEKHWNV 356
+ T + F D + FS+ +LR A + K +T K +++Q+ H+ V
Sbjct 279 NFTRVNFGVDTDSSEG-------DFSVSSLRAAFAVDKLLSVTMRAGKTFQDQMRAHYGV 331
Query 357 SPGDGFSEMCTYLGGISSSLDINEVVNQNITGSNAAD----------IAGKGTGVSNGVI 406
D YLGG S + +++V +G+ A + +AGKGTG G I
Sbjct 332 EIPDSRDGRVNYLGGFDSDMQVSDVTQ--TSGTTATEYKPEAGYLGRVAGKGTGSGRGRI 389
Query 407 NFNSQGRYGVVMCIYHCLPLIDYTTDFVSPSVTRVNAADFAIPEFDRVGMQTVPLSYVSN 466
F+++ +GV+MCIY +P I Y + P V +++ D+ PEF+ +GMQ + SY+S
Sbjct 390 VFDAK-EHGVLMCIYSLVPQIQYDCTRLDPMVDKLDRFDYFTPEFENLGMQPLNSSYIS- 447
Query 467 GPLSVLPLSIPNEI-GYAPRYIDYKTDIDTSIGAFKTS--LKNWVISYDNQSLANQFGYS 523
S N + GY PRY +YKT +D + G F S L +W +S +F
Sbjct 448 ---SFCTTDPKNPVLGYQPRYSEYKTALDVNHGQFAQSDALSSWSVS--------RFRRW 496
Query 524 VETPESPVPNPANSSWNYTLFKVNPNSLNPLFAVEADSSIDTDQFLCSTFFDVKVVRNLD 583
P+ + + FK++P LN +F V+ + + D F++ V ++
Sbjct 497 TTFPQLEIAD----------FKIDPGCLNSIFPVDYNGTEANDCVYGGCNFNIVKVSDMS 546
Query 584 TDGLP 588
DG+P
Sbjct 547 VDGMP 551
>gi|496521299|ref|WP_009229582.1| capsid protein [Prevotella sp. oral taxon 317]
gi|288330570|gb|EFC69154.1| putative capsid protein (F protein) [Prevotella sp. oral taxon
317 str. F0108]
Length=541
Score = 223 bits (568), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 180/606 (30%), Positives = 284/606 (47%), Gaps = 87/606 (14%)
Query 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEVLPGDTFKIDLKSFTRTQP 60
+ + +K R R+ FDLS K +TA AG LLPV + +++ D +I + F RT P
Sbjct 3 LKKVPQIKPSRANRPRSAFDLSQKHLYTAPAGALLPVLSVDLMFHDHIRIQAQDFMRTMP 62
Query 61 LNTAAFARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHALDSSPLNVVKLDGSMPFTD 120
+N+AAF MR Y+F+FVPY LW+ + +T M D + ++ SS LD S+P
Sbjct 63 MNSAAFISMRGVYEFFFVPYSQLWHPYDQFITSMNDY-RSSVVSSAAGDKALD-SVPNVK 120
Query 121 LSSISRYLNSLASNSTAVTNKANYFGYNRALCSAKLMECLGYGNLYYYAESTSNTFAKRP 180
L+ + +++ T+K + FGY + S +LM+ LGYG + +++ P
Sbjct 121 LADMYKFVRER-------TDK-DIFGYPHSNNSCRLMDLLGYG------KPITSSKTPVP 166
Query 181 LHYNLNVSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYMPYSTGSSGMMLSFNYSDFY 240
L Y NV+LF LLAY KIY+DYYR++ +E V FN+D+ G + +D +
Sbjct 167 LLYTGNVNLFRLLAYNKIYSDYYRNTTYEGVDVYSFNIDH------KKGTFVP--TADEF 218
Query 241 ENYSMFDLRYCNWQKDLFHGVVPNQQY---GDVASISMSVPVVAGSSAAlinsritssns 297
+ Y +L Y N D + + P + D S + + GS+ + N
Sbjct 219 KKY--LNLHYRNAPLDFYTNLRPTPLFTIGSDSFSSVLQLSDPTGSAGFSADGNSAKLNM 276
Query 298 tttlrFPTDPAIPDATPLLTHPSFSILALRQAEFLQKWKEITQSGNKDYKEQVEKHWNVS 357
A PD ++ A+R A L K I+ K Y EQ+E H+ V+
Sbjct 277 ----------ASPDV--------LNVSAIRSAFALDKLLSISMRAGKTYAEQIEAHFGVT 318
Query 358 PGDGFSEMCTYLGGISSSLDINEV------VNQNITGSNAADIA-------GKGTGVSNG 404
+G YLGG S++ + +V N N++ A +A GKGTG G
Sbjct 319 VSEGRDGQVYYLGGFDSNVQVGDVTQTSGTTNPNVSEVGNAKLAGYLGKITGKGTGSGYG 378
Query 405 VINFNSQGRYGVVMCIYHCLPLIDYTTDFVSPSVTRVNAADFAIPEFDRVGMQTVPLSYV 464
I F+++ GV+MCIY +P + Y + P V + D+ IPEF+ +GMQ + ++V
Sbjct 379 EIQFDAK-EPGVLMCIYSVVPAMQYDCMRLDPFVAKQTRGDYFIPEFENLGMQPIVPAFV 437
Query 465 SNGPLSVLPLSIPNEIGYAPRYIDYKTDIDTSIGAFKTS--LKNWVISYDNQSLANQFGY 522
S L + N G+ PRY +YKT D + G F L W I+ S
Sbjct 438 S------LNRAKDNSYGWQPRYSEYKTAFDINHGQFANGEPLSYWSIARARGS------- 484
Query 523 SVETPESPVPNPANSSWNYTLFKVNPNSLNPLFAVEADSSIDTDQFLCSTFFDVKVVRNL 582
+++N K+NP+ L+ +FAV + + TD F+++ V ++
Sbjct 485 -----------DTLNTFNVAALKINPHWLDSVFAVNYNGTEVTDCMFGYAHFNIEKVSDM 533
Query 583 DTDGLP 588
DG+P
Sbjct 534 TEDGMP 539
>gi|490477384|ref|WP_004347761.1| capsid protein [Prevotella buccalis]
gi|281300712|gb|EFA93043.1| putative capsid protein (F protein) [Prevotella buccalis ATCC
35310]
Length=552
Score = 219 bits (557), Expect = 1e-59, Method: Compositional matrix adjust.
Identities = 174/608 (29%), Positives = 274/608 (45%), Gaps = 91/608 (15%)
Query 7 LKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEVLPGDTFKIDLKSFTRTQPLNTAAF 66
+K+ R RN FDLS K FTA AG LLPV T +++P D I F R P+N+AAF
Sbjct 8 IKASRANRPRNAFDLSQKHLFTAHAGMLLPVMTLDLIPHDHVSIQATDFMRCLPMNSAAF 67
Query 67 ARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHALDSSPLNVVKLDGSMPFTDLSSISR 126
MR Y+F+FVPY LW+ + +T M N ++ S L K +P +
Sbjct 68 MSMRSVYEFFFVPYSQLWHPFDQFITGM--NDYRSVLQSDLYKSKSPLVIPSFKRKELYE 125
Query 127 YLNSLASNSTAVTNKANYFGYNRALCSAKLMECLGYGNLYYYAESTSNTFA-KRPLHYNL 185
N+ +N+ + FG+ +L++ LGYG +Y A+ +S A + L
Sbjct 126 LFNAPGGFLNQQSNQPDIFGFKSRFNFLRLLDLLGYG-VYVNADGSSRIDAFSKLLDDTE 184
Query 186 NVSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYMPYSTGSSGMMLSFNYSDFYENYSM 245
+S+F L AYQKIY+D+YR++ +E V S F++D + S + F
Sbjct 185 KLSIFRLAAYQKIYSDFYRNTTYEAVDVSSFSLDNITDSISAINAFKRFG---------- 234
Query 246 FDLRYCNWQKDLFHGVVPNQQYGDVASISM-----------SVPVVAGSSAAlinsrits 294
LRY N Q D F + P + D+ + S+ SV + + S+A
Sbjct 235 -TLRYRNAQLDYFTNLRPTPLF-DLDNPSLNSFYNTPGNADSVSIDSDSNAV-------- 284
Query 295 snstttlrFPTDPAIPDATPLLTHPSFSILALRQAEFLQKWKEITQSGNKDYKEQVEKHW 354
F D + LLT + ++R A L K ITQ K Y EQ++ H+
Sbjct 285 -------NFQLD------SDLLT-----VQSIRNAFALDKLMRITQRAGKTYAEQIKAHF 326
Query 355 NVSPGDGFSEMCTYLGGISSSLDINEVVNQNIT------------GSNAADIAGKGTGVS 402
+G Y+GG S++ + +V + T G + GK G
Sbjct 327 GFEVSEGRDGRVNYIGGFDSNIQVGDVTQMSGTTASPEQGVSIKHGGYLGRVTGKAQGSG 386
Query 403 NGVINFNSQGRYGVVMCIYHCLPLIDYTTDFVSPSVTRVNAADFAIPEFDRVGMQTVPLS 462
+G I F++ +G++MCIY +P + Y + P VT+++ DF +PEF+ +GMQ +
Sbjct 387 SGHIEFDAH-EHGILMCIYSLVPDMQYDATRIDPFVTKLSRGDFFMPEFEDLGMQPLQTR 445
Query 463 YVSNGPLSVLPLSIPNEIGYAPRYIDYKTDIDTSIGAFKTS--LKNWVISYDNQSLANQF 520
Y+S+ + G+ PRY +YKT +D + G F L W +
Sbjct 446 YISD-----IRTQTEKFKGWQPRYSEYKTSLDINHGQFANGQPLSYWTVGR--------- 491
Query 521 GYSVETPESPVPNPANSSWNYTLFKVNPNSLNPLFAVEADSSIDTDQFLCSTFFDVKVVR 580
G + ET E +++ K+NP L+ +FAV + + TD F+V+ V
Sbjct 492 GRAGETLE---------TFDIASLKINPKWLDSIFAVNYNGTQITDCVFGGCQFNVQKVS 542
Query 581 NLDTDGLP 588
++ +G P
Sbjct 543 DMSENGEP 550
>gi|517172762|ref|WP_018361580.1| hypothetical protein [Prevotella nanceiensis]
Length=568
Score = 219 bits (557), Expect = 1e-59, Method: Compositional matrix adjust.
Identities = 184/613 (30%), Positives = 281/613 (46%), Gaps = 86/613 (14%)
Query 7 LKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEVLPGDTFKIDLKSFTRTQPLNTAAF 66
+K + RN FD+S + FTA AG LLPV + ++LP D +I+ F RT P+N+AAF
Sbjct 9 IKPSKATRPRNAFDISQRHLFTAPAGALLPVLSLDLLPHDHVEINASDFMRTLPMNSAAF 68
Query 67 ARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHALDSSPLNVVKLDGSMPFTDLSSISR 126
MR Y+FYFVPY LW+ + +T M D SS + K G P + +S +
Sbjct 69 MSMRGVYEFYFVPYKQLWSGFDQFITGMSD-----YKSSFMYAFK--GKTPPSCVSFDVQ 121
Query 127 YLNSLASNSTAVTNKANYFGYNRALCSAKLMECLGYGNLYY-----YAESTSNTFAKRPL 181
L +TA + G+++ ++++ LGYG Y TS T K
Sbjct 122 KLVDWCKTNTA----KDIHGFDKNKGVYRILDLLGYGKYANSAGVPYTNPTSTTMGK--- 174
Query 182 HYNLNVSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYMPYSTGSSGMMLSFNYSDFYE 241
+ F LAYQKIY D+YR++ +E FNVD M Y +G + D
Sbjct 175 -----CTPFRGLAYQKIYNDFYRNTTYEEYQLESFNVD-MFYGSGKVKETIPNEPWD--- 225
Query 242 NYSMFDLRYCNWQKDLFHGVVP---------NQQY--GDVASISMSVPVVAGSSAAlins 290
Y F LRY N QKDL V P N Q+ G + P V G + +S
Sbjct 226 -YDWFTLRYRNAQKDLLTNVRPTPLFSIDDFNPQFFTGGSDIVMEKGPNVTGGTHEYRDS 284
Query 291 ritssnstttlrFPTDPAIPDATPLLTHPSFSILALRQAEFLQKWKEITQSGNKDYKEQV 350
+ + + + S+ +R A L+K +T K YKEQ+
Sbjct 285 VVIVGKNLKENGVDSKRTM-----------ISVADIRNAFALEKLASVTMRAGKTYKEQM 333
Query 351 EKHWNVSPGDGFSEMCTYLGGISSSLDINEVVNQNIT----------GSNAADIAGKGTG 400
E H+ +S +G CTY+GG S++ + +V + T G GK TG
Sbjct 334 EAHFGISVEEGRDGRCTYIGGFDSNIQVGDVTQSSGTTVTGTKDTSFGGYLGRTTGKATG 393
Query 401 VSNGVINFNSQGRYGVVMCIYHCLPLIDYTTDFVSPSVTRVNAADFAIPEFDRVGMQTV- 459
+G I F+++ +G++MCIY +P + Y + V P V ++ DF +PEF+ +GMQ +
Sbjct 394 SGSGHIRFDAK-EHGILMCIYSLVPDVQYDSKRVDPFVQKIERGDFFVPEFENLGMQPLF 452
Query 460 --PLSYVSNGPLSVLPLSIPNEIGYAPRYIDYKTDIDTSIGAF--KTSLKNWVISYDNQS 515
+SY N + + G+ PRY +YKT +D + G F + L W +
Sbjct 453 AKNISYKYNNNTANSRIKNLGAFGWQPRYSEYKTALDINHGQFVHQEPLSYWTV------ 506
Query 516 LANQFGYSVETPESPVPNPANSSWNYTLFKVNPNSLNPLFAVEADSSIDTDQFLCSTFFD 575
A G S+ S++N + FK+NP L+ +FAV + + TDQ +F+
Sbjct 507 -ARARGESM------------SNFNISTFKINPKWLDDVFAVNYNGTELTDQVFGGCYFN 553
Query 576 VKVVRNLDTDGLP 588
+ V ++ DG+P
Sbjct 554 IVKVSDMSIDGMP 566
Lambda K H a alpha
0.317 0.133 0.401 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4356774862695