bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-12_CDS_annotation_glimmer3.pl_2_6
Length=50
Score E
Sequences producing significant alignments: (Bits) Value
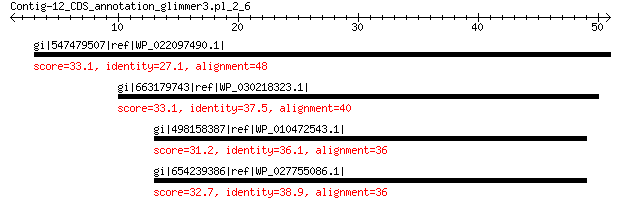
gi|547479507|ref|WP_022097490.1| putative uncharacterized protein 33.1 3.9
gi|663179743|ref|WP_030218323.1| DNA-binding protein 33.1 5.9
gi|498158387|ref|WP_010472543.1| DNA-binding protein 31.2 7.3
gi|654239386|ref|WP_027755086.1| XRE family transcriptional regu... 32.7 9.9
>gi|547479507|ref|WP_022097490.1| putative uncharacterized protein [Eubacterium eligens CAG:72]
gi|524278470|emb|CDA41448.1| putative uncharacterized protein [Eubacterium eligens CAG:72]
Length=148
Score = 33.1 bits (74), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 13/48 (27%), Positives = 31/48 (65%), Gaps = 0/48 (0%)
Query 3 KQFYLVLLHYGSEVSSKVVISLDRLDSFLSRGYTVEFVQPIRVLSDEL 50
+ + + L YG++ + +V+ DR+D+ + G++ E VQ + V++D++
Sbjct 87 RSWNVSLTDYGAQKADEVIRIFDRIDNVIYNGFSEEEVQTLSVMADKV 134
>gi|663179743|ref|WP_030218323.1| DNA-binding protein [Streptomyces griseoluteus]
Length=235
Score = 33.1 bits (74), Expect = 5.9, Method: Composition-based stats.
Identities = 15/40 (38%), Positives = 23/40 (58%), Gaps = 0/40 (0%)
Query 10 LHYGSEVSSKVVISLDRLDSFLSRGYTVEFVQPIRVLSDE 49
L S ++V DR+D S+G++V V P+R +SDE
Sbjct 146 LARASASGAEVAFEEDRMDEAFSQGWSVLVVGPVRTVSDE 185
>gi|498158387|ref|WP_010472543.1| DNA-binding protein, partial [Streptomyces somaliensis]
Length=85
Score = 31.2 bits (69), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 20/36 (56%), Gaps = 0/36 (0%)
Query 13 GSEVSSKVVISLDRLDSFLSRGYTVEFVQPIRVLSD 48
G +V +D +D SRG++V V P RV++D
Sbjct 2 GRAAGREVAFEVDHIDEAFSRGWSVLLVGPARVVTD 37
>gi|654239386|ref|WP_027755086.1| XRE family transcriptional regulator [Streptomyces sp. CNH099]
Length=309
Score = 32.7 bits (73), Expect = 9.9, Method: Composition-based stats.
Identities = 14/36 (39%), Positives = 23/36 (64%), Gaps = 0/36 (0%)
Query 13 GSEVSSKVVISLDRLDSFLSRGYTVEFVQPIRVLSD 48
+ V S+V +D +D +SRG++V V P RV++D
Sbjct 232 AAAVGSEVAFEVDHVDEAMSRGWSVLAVGPARVVTD 267
Lambda K H a alpha
0.323 0.140 0.378 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 443773169100