bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-12_CDS_annotation_glimmer3.pl_2_3
Length=612
Score E
Sequences producing significant alignments: (Bits) Value
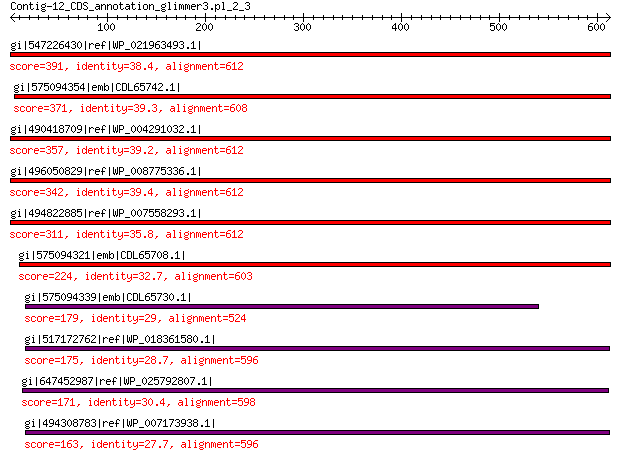
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 391 2e-124
gi|575094354|emb|CDL65742.1| unnamed protein product 371 3e-116
gi|490418709|ref|WP_004291032.1| hypothetical protein 357 3e-111
gi|496050829|ref|WP_008775336.1| hypothetical protein 342 2e-105
gi|494822885|ref|WP_007558293.1| hypothetical protein 311 2e-93
gi|575094321|emb|CDL65708.1| unnamed protein product 224 1e-60
gi|575094339|emb|CDL65730.1| unnamed protein product 179 3e-45
gi|517172762|ref|WP_018361580.1| hypothetical protein 175 5e-44
gi|647452987|ref|WP_025792807.1| hypothetical protein 171 2e-42
gi|494308783|ref|WP_007173938.1| hypothetical protein 163 6e-40
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 391 bits (1004), Expect = 2e-124, Method: Compositional matrix adjust.
Identities = 235/631 (37%), Positives = 341/631 (54%), Gaps = 77/631 (12%)
Query 1 MAGLFSYGDIKNKPRRSGFDLGNKNAFTAKVGELLPVYWKFCLPGDKFKISQEWFTRTQP 60
M+ + S +KN +R+GFDL KNAFTAKVGELLP+ K PGDKF I + FTRTQP
Sbjct 1 MSSVMSLTALKNSVKRNGFDLSFKNAFTAKVGELLPIMCKEVYPGDKFNIRGQAFTRTQP 60
Query 61 VDTSAFTRIREYYEWFFVPLHLMYRNSNEAIMSMENQPNYAASGTQSITFNRKLPWVDLL 120
V+++A++R+REYY+++FVP L++ + +M + P++AA S+ +++ PW
Sbjct 61 VNSAAYSRLREYYDFYFVPYRLLWNMAPTFFTNMPD-PHHAADLVSSVNLSQRHPWFTFF 119
Query 121 TLNNAVENVKA-----STYHDNMFGFSRALGFAKLYNYLGVG------QVDPSKTLANLR 169
+ + N+ + Y N FGFSR KL NYL G V ++
Sbjct 120 DIMEYLGNLNSLSGAYEKYQKNFFGFSRVELSVKLLNYLNYGFGKDYESVKVPSDSDDIV 179
Query 170 ISAFPFYAYQKIYNDYYRNSQWEVNKPWTYNCDFWNGEDT---TPVASSKDLFDTNPNDS 226
+S FP AYQKI DY+R+ QW+ P+ YN D+ G+ + P++S + D N +
Sbjct 180 LSPFPLLAYQKICEDYFRDDQWQSAAPYRYNLDYLYGKSSGFHIPMSSFTN--DAFKNPT 237
Query 227 IFELRYANWNKDLYMGAMPNAQFGDVAFVPVDSSGKLPVSLPSIEVGGVAPIYNTGAGGV 286
+F+L Y N+ KD + G +P AQ+GDV+ +PI+
Sbjct 238 MFDLNYCNFQKDYFTGMLPRAQYGDVSVA--------------------SPIFG------ 271
Query 287 QPDAQIGLRGAIT--GAPDNGQTVTAYGADKTDAARPYFYAVPDGSVAHLKTNAKTIQVP 344
D IG ++T AP G G V + N+ T
Sbjct 272 --DLDIGDSSSLTFASAPQQGANTIQSG------------------VLVVNNNSNT---- 307
Query 345 YEFSSKFDVLQLRAAECLQKWKEIAQANGQNYASQVKAHFGVSPNPMTSHRCQRVCGFDG 404
++ VL LR AECLQKW+EIAQ+ +Y +Q++ HF VSP+ S C+ + G+
Sbjct 308 ---TAGLSVLALRQAECLQKWREIAQSGKMDYQTQMQKHFNVSPSATLSGHCKYLGGWTS 364
Query 405 SIDISAVENTNLSSD-EAIIRGKGIGGYRVNKPETFKTTEHGVLMCIYHAVPLLDYAPTG 463
++DIS V NTNL+ D +A I+GKG G NK + F+++EHG++MCIYH +PLLD++
Sbjct 365 NLDISEVVNTNLTGDNQADIQGKGTGTLNGNKVD-FESSEHGIIMCIYHCLPLLDWSINR 423
Query 464 PDLQFMTTVDGDSWPVPELDSVGFEEL-PSYSLLNTSDVQPIKEPRPFGYVPRYISWKTS 522
Q T D + +PE DSVG ++L PS + D+ GYVPRY KTS
Sbjct 424 IARQNFKTTFTD-YAIPEFDSVGMQQLYPSEMIFGLEDLPSDPSSINMGYVPRYADLKTS 482
Query 523 VDVVRGAFIDTLKSWTAPIGEDYMKIYFDNNNVPGGAHFGF-YTWFKVNPSVVNPIFGVV 581
+D + G+FIDTL SW +P+ + Y+ Y G + Y +FKVNP +V+ IFGV
Sbjct 483 IDEIHGSFIDTLVSWVSPLTDSYISAYRQACKDAGFSDITMTYNFFKVNPHIVDNIFGVK 542
Query 582 ADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 612
AD + NTDQLL+N FD++ RN Y+GLPY
Sbjct 543 ADSTINTDQLLINSYFDIKAVRNFDYNGLPY 573
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 371 bits (953), Expect = 3e-116, Method: Compositional matrix adjust.
Identities = 239/637 (38%), Positives = 347/637 (54%), Gaps = 51/637 (8%)
Query 5 FSYGDIKNKPRRSGFDLGNKNAFTAKVGELLPVYWKFCLPGDKFKISQEWFTRTQPVDTS 64
S DIKN+P R+GFDL K FTAK GELLPV K LPGD F I+ FTRTQP++TS
Sbjct 1 MSMADIKNRPSRNGFDLSFKKNFTAKAGELLPVMTKVVLPGDSFNINLRSFTRTQPLNTS 60
Query 65 AFTRIREYYEWFFVPLHLMYRNSNEAIMSMENQPNYAASGT--QSITFNRKLPWVDLLTL 122
AF R+REYY+++FVP M+ + I M +A+ T + + ++P+ +
Sbjct 61 AFARMREYYDFYFVPFEQMWNKFDSCITQMNANVQHASGPTLDDNTPLSGRMPYFTSEQI 120
Query 123 NNAVENVKASTYHDNMFGFSRALGFAKLYNYLGVGQVDP--SKT--------LANLRISA 172
+ + N +A+ N FGF+R+ KL YLG G + S+T L NL +S
Sbjct 121 ADYL-NDQATAARKNPFGFNRSTLTCKLLQYLGYGDYNSFDSETNTWSAKPLLYNLELSP 179
Query 173 FPFYAYQKIYNDYYRNSQWEVNKPWTYNCDFWNGEDTTPVASSKDLFDTNPNDSIFELRY 232
FP AYQKIY+D+YR +QWE P T+N D+ G + + D N + F++RY
Sbjct 180 FPLLAYQKIYSDFYRYTQWEKTNPSTFNLDYIKGTSDLQMDLTGLPSDDN---NFFDIRY 236
Query 233 ANWNKDLYMGAMPNAQFGDVAFVPVDSSGKLPVSLPSIEVGGVAPIYNTGA--GGVQPDA 290
N+ KD++ G +P AQ+G + VP++ G+L V I G PI+ T G +
Sbjct 237 CNYQKDMFHGVLPVAQYGSASVVPIN--GQLNV----ISNGDSGPIFKTSTPDPGTPGTS 290
Query 291 QIGLRGAITGAPDNGQTVTAYGADKTDAARPYFYAVP-DGSVAHLKTNAKTIQVPYEFSS 349
+ + G I G + V+ + +A P Y P + S L + +
Sbjct 291 YVTVGGNI-GVDNRSFGVSGSTLNVGKSADPSGYGFPSNASTRSLLWENPNLIIENNQGF 349
Query 350 KFDVLQLRAAECLQKWKEIAQANGQNYASQVKAHFGVSPNPMTSHRCQRVCGFDGSIDIS 409
+L LR AE LQKWKE++ + ++Y SQ++ H+G+ + SH+ + + G S+DI+
Sbjct 350 YVPILALRQAEFLQKWKEVSVSGEEDYKSQIEKHWGIKVSDFLSHQARYLGGCATSLDIN 409
Query 410 AVENTNLSSDEAI-IRGKGIGGYRVNKPETFKTT-EHGVLMCIYHAVPLLDYAPTGPDLQ 467
V N N++ D A I GKG + N F++ E+G++MCIYH +P++DY +G D
Sbjct 410 EVINNNITGDNAADIAGKGT--FTGNGSIRFESKGEYGIIMCIYHVLPIVDYVGSGVD-H 466
Query 468 FMTTVDGDSWPVPELDSVGFEELPSYSLLNTSDVQPIKEPRP------FGYVPRYISWKT 521
T VD S+P+PELD +G E +P +N P+KE GY PRYI WKT
Sbjct 467 SCTLVDATSFPIPELDQIGMESVPLVRAMN-----PVKESDTPSADTFLGYAPRYIDWKT 521
Query 522 SVDVVRGAFIDTLKSWTAPIGEDYM----KIYFDNN-NV-PGGAHFGFYTWFKVNPSVVN 575
SVD G F D+L++W P+G+ + + F +N NV P GF FKVNPS+V+
Sbjct 522 SVDRSVGDFADSLRTWCLPVGDKELTSANSLNFPSNPNVEPDSIAAGF---FKVNPSIVD 578
Query 576 PIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 612
P+F VVAD + TD+ L + FDV+V RNL +GLPY
Sbjct 579 PLFAVVADSTVKTDEFLCSSFFDVKVVRNLDVNGLPY 615
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 357 bits (916), Expect = 3e-111, Method: Compositional matrix adjust.
Identities = 240/640 (38%), Positives = 324/640 (51%), Gaps = 90/640 (14%)
Query 1 MAGLFSYGDIKNKPRRSGFDLGNKNAFTAKVGELLPVYWKFCLPGDKFKISQEWFTRTQP 60
MA + S I+NKP R+GFDL K FTAK GELLPV K LPGD FKI+ + FTRTQP
Sbjct 1 MANIMSLKSIRNKPSRNGFDLSFKKNFTAKAGELLPVMVKEVLPGDTFKINLKAFTRTQP 60
Query 61 VDTSAFTRIREYYEWFFVPLHLMYRNSNEAIMSMENQPNYAAS--GTQSITFNRKLPWVD 118
V+T+AF RIREYY++FFVP L++ +N + M + P +A S T++ + ++P++
Sbjct 61 VNTAAFARIREYYDFFFVPYDLLWNKANTVLTQMYDNPQHAVSIDPTRNFVLSGEMPYMT 120
Query 119 ---LLTLNNAVENVKA-STYHDNMFGFSRALGFAKLYNYLGVGQVDPSKT--------LA 166
+ + NA+ A + Y N FG++R+ KL YLG G + T +A
Sbjct 121 SEAIASYINALSTASALADYKSNYFGYNRSKSSVKLLEYLGYGNYESFLTDDWNTAPLMA 180
Query 167 NLRISAFPFYAYQKIYNDYYRNSQWEVNKPWTYNCDFWNGEDTTPVASSKDLFDTNPNDS 226
NL + F AYQKIY+D+YR+SQWE P T+N D+ +G + F N N
Sbjct 181 NLNHNIFGLLAYQKIYSDFYRDSQWERVSPSTFNVDYLDGSSMNLDNAYSTEFYQNYN-- 238
Query 227 IFELRYANWNKDLYMGAMPNAQFGDVAFVPV--DSSGKLPVSLPSIEVGGVAPIYNTGAG 284
F+LRY NW KDL+ G +P+ Q+G+ A + D +GKL +S N
Sbjct 239 FFDLRYCNWQKDLFHGVLPHQQYGETAVASITPDVTGKLTLS-------------NFSTV 285
Query 285 GVQPDAQIGLRGAITGAPDNGQTVTAYGADKTDAARPYFYAVPDGSVAHLKTNAKTIQVP 344
G P TA G + P F V D S
Sbjct 286 GTSP-------------------TTASGTATKNL--PAFDTVGDLS-------------- 310
Query 345 YEFSSKFDVLQLRAAECLQKWKEIAQANGQNYASQVKAHFGVSPNPMTSHRCQRVCGFDG 404
+L LR AE LQKWKEI Q+ ++Y Q++ H+GVS S C + G
Sbjct 311 --------ILVLRQAEFLQKWKEITQSGNKDYKDQLEKHWGVSVGDGFSELCTYLGGVSS 362
Query 405 SIDISAVENTNLS-SDEAIIRGKGIGGYRVNKPETFKTT-EHGVLMCIYHAVPLLDYAPT 462
SIDI+ V NTN++ S A I GKG+G N F + +G++MCIYH +PLLDY
Sbjct 363 SIDINEVINTNITGSAAADIAGKGVG--VANGEINFNSNGRYGLIMCIYHCLPLLDYTTD 420
Query 463 GPDLQFMTTVDGDSWPVPELDSVGFEELPSYSLLNTSDVQPIKEPRPFGYVPRYISWKTS 522
D F+ V+ + +PE D VG + +P L+N GYVPRYI +KTS
Sbjct 421 MLDPAFL-KVNSTDYAIPEFDRVGMQSMPLVQLMNPLRSFANASGLVLGYVPRYIDYKTS 479
Query 523 VDVVRGAFIDTLKSWTAPIGEDYM--KIYFDNNN--------VPGGAHFGFYTWFKVNPS 572
VD G F TL SW G + ++ N+ VP A F T+FKVNP
Sbjct 480 VDQSVGGFKRTLNSWVISYGNISVLKQVTLPNDAPPIEPSEPVPSVAPMNF-TFFKVNPD 538
Query 573 VVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 612
++PIF V A NTDQ L + FD++ RNL DGLPY
Sbjct 539 CLDPIFAVQAGDDTNTDQFLCSSFFDIKAVRNLDTDGLPY 578
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 342 bits (877), Expect = 2e-105, Method: Compositional matrix adjust.
Identities = 241/649 (37%), Positives = 325/649 (50%), Gaps = 106/649 (16%)
Query 1 MAGLFSYGDIKNKPRRSGFDLGNKNAFTAKVGELLPVYWKFCLPGDKFKISQEWFTRTQP 60
MA + S ++NK R+GFDL +K FTAK GELLPV LPGDK+ I + FTRTQP
Sbjct 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKPGELLPVKCWEVLPGDKWSIDLKSFTRTQP 60
Query 61 VDTSAFTRIREYYEWFFVPLHLMYRNSNEAIMSMENQPNYAASGTQSITFNRKLPWV--- 117
++T+AF R+REYY+++FVP +L++ +N + M + P +A S S N+ L V
Sbjct 61 LNTAAFARMREYYDFYFVPYNLLWNKANTVLTQMYDNPQHATSYIPSA--NQALAGVMPN 118
Query 118 -------DLLTLNNAVENVKASTYHDNMFGFSRALGFAKLYNYLGVGQV----------- 159
D L L A + ++Y N FG+SR+LG AKL YLG G
Sbjct 119 VTCKGIADYLNL-VAPDVTTTNSYEKNYFGYSRSLGTAKLLEYLGYGNFYTYATSKNNTW 177
Query 160 DPSKTLANLRISAFPFYAYQKIYNDYYRNSQWEVNKPWTYNCDFWNGEDTTPVASSKDLF 219
S +NL+++ + AYQKIY D+ R+SQWE P +N D+ +G T A + D
Sbjct 178 TKSPLSSNLQLNIYGVLAYQKIYADHIRDSQWEKVSPSCFNVDYLSG--TVDSAMTIDSM 235
Query 220 DTN----PNDSIFELRYANWNKDLYMGAMPNAQFGDVAFVPVDSSGKLP----VSLPSIE 271
T P ++F+LRY NW KDL+ G +P Q+GD A V V+ S L V P +
Sbjct 236 ITGQGFAPFYNMFDLRYCNWQKDLFHGVLPRQQYGDTAAVNVNLSNVLSAQYMVQTPDGD 295
Query 272 VGGVAPIYNTGAGGVQPDAQIGLRGAITGAPDNGQTVTAYGADKTDAARPYFYAVPDGSV 331
G +P +TG N QTV G
Sbjct 296 PVGGSPFSSTGV--------------------NLQTVNGSGT------------------ 317
Query 332 AHLKTNAKTIQVPYEFSSKFDVLQLRAAECLQKWKEIAQANGQNYASQVKAHFGVSPNPM 391
F VL LR AE LQKWKEI Q+ ++Y Q++ H+ VS
Sbjct 318 -------------------FTVLALRQAEFLQKWKEITQSGNKDYKDQIEKHWNVSVGEA 358
Query 392 TSHRCQRVCGFDGSIDISAVENTNLS-SDEAIIRGKG--IGGYRVNKPETFKTTE-HGVL 447
S + G S+DI+ V N N++ S+ A I GKG +G R+ +F E +G++
Sbjct 359 YSEMSLYLGGTTASLDINEVVNNNITGSNAADIAGKGVVVGNGRI----SFDAGERYGLI 414
Query 448 MCIYHAVPLLDYAPTGPDLQFMTTVDGDSWPVPELDSVGFEELPSYSLLNTSDVQPIKEP 507
MCIYH++PLLDY + F T ++ + +PE D VG E +P SL+N
Sbjct 415 MCIYHSLPLLDYTTDLVNPAF-TKINSTDFAIPEFDRVGMESVPLVSLMNPLQSSYNVGS 473
Query 508 RPFGYVPRYISWKTSVDVVRGAFIDTLKSWTAPIGE----DYMKIYFDNNNVPGGAHFGF 563
GY PRYIS+KT VD GAF TLKSW + + D NN PG
Sbjct 474 SILGYAPRYISYKTDVDSSVGAFKTTLKSWVMSYDNQSVINQLNYQDDPNNSPG--TLVN 531
Query 564 YTWFKVNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 612
YT FKVNP+ V+P+F V A S +TDQ L + FDV+V RNL DGLPY
Sbjct 532 YTNFKVNPNCVDPLFAVAASNSIDTDQFLCSSFFDVKVVRNLDTDGLPY 580
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 311 bits (798), Expect = 2e-93, Method: Compositional matrix adjust.
Identities = 219/647 (34%), Positives = 328/647 (51%), Gaps = 76/647 (12%)
Query 1 MAGLFSYGDIKNKPRRSGFDLGNKNAFTAKVGELLPVYWKFCLPGDKFKISQEWFTRTQP 60
MA + S ++NKP R+G+DL K FTAK G L+PV+W LP D + + F RTQP
Sbjct 8 MANIMSMKSVRNKPTRAGYDLTQKINFTAKAGSLIPVWWTPVLPFDDLNATVKSFVRTQP 67
Query 61 VDTSAFTRIREYYEWFFVPLHLMYRNSNEAIMSMENQPNYAASGT--QSITFNRKLPWVD 118
++T+AF R+R Y++++FVP M+ AI M +A+ ++ + +LP+
Sbjct 68 LNTAAFARMRGYFDFYFVPFRQMWNKFPTAITQMRTNLLHASGPVLADNVPLSDELPY-- 125
Query 119 LLTLNNAVENVKASTYHDNMFGFSRALGFAKLYNYLGVGQVDP---------------SK 163
T + + + N FG+ RA + YLG G P
Sbjct 126 -FTAEQVADYIVSLADSKNQFGYYRAWLVCIILEYLGYGDFYPYIVEAAGGEGATWATRP 184
Query 164 TLANLRISAFPFYAYQKIYNDYYRNSQWEVNKPWTYNCDFWNGE-DTTPVASSKDLFDTN 222
L NL+ S FP +AYQKIY D+ R +QWE + P T+N D+ +G D+ + + + F +
Sbjct 185 MLNNLKFSPFPLFAYQKIYADFNRYTQWERSNPSTFNIDYISGSADSLQLDFTVEGFKDS 244
Query 223 PNDSIFELRYANWNKDLYMGAMPNAQFGDVAFVPVDSSGKLPVSLPSIEVGGVAPIYNTG 282
N +F++RY+NW +DL G +P AQ+G+ + VPV SG + V +E G P + TG
Sbjct 245 FN--LFDMRYSNWQRDLLHGTIPQAQYGEASAVPV--SGSMQV----VE-GPTPPAFTTG 295
Query 283 AGGVQPDAQIGLRGAITGAPDNGQTVTAYGADKTDAARPYFYAVPDGSVAHLKTNAKTIQ 342
GV A + I G+ Q T+ G + L+ N
Sbjct 296 QDGV---AFLNGNVTIQGSSGYLQAQTSVGESRI-----------------LRFNNTNSG 335
Query 343 VPYEFSSKFDV--LQLRAAECLQKWKEIAQANGQNYASQVKAHFGVSPNPMTSHRCQRVC 400
+ E S F V L LR AE QKWKE+A A+ ++Y SQ++AH+G S N S CQ +
Sbjct 336 LIVEGDSSFGVSILALRRAEAAQKWKEVALASEEDYPSQIEAHWGQSVNKAYSDMCQWLG 395
Query 401 GFDGSIDISAVENTNLSSDEAI-IRGKGIGGYRVNKPETFKT-TEHGVLMCIYHAVPLLD 458
+ + I+ V N N++ + A I GKG N F ++G++MC++H +P LD
Sbjct 396 SINIDLSINEVVNNNITGENAADIAGKGT--MSGNGSINFNVGGQYGIVMCVFHVLPQLD 453
Query 459 YAPTGPDLQFMTTVDGD-SWPVPELDSVGFEELPSYSLLNTSDVQP------IKEPRPFG 511
Y + P F TT+ +P+PE D +G E++P LN V+P + FG
Sbjct 454 YITSAP--HFGTTLTNVLDFPIPEFDKIGMEQVPVIRGLNP--VKPKDGDFKVSPNLYFG 509
Query 512 YVPRYISWKTSVDVVRGAFIDTLKSWTAPIGEDYMKI-----YFDNNNVPG-GAHFGFYT 565
Y P+Y +WKT++D G F +LK+W P ++ + + DN NV GF
Sbjct 510 YAPQYYNWKTTLDKSMGEFRRSLKTWIIPFDDEALLAADSVDFPDNPNVEADSVKAGF-- 567
Query 566 WFKVNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 612
FKV+PSV++ +F V A+ NTDQ L + FDV V R+L +GLPY
Sbjct 568 -FKVSPSVLDNLFAVKANSDLNTDQFLCSTLFDVNVVRSLDPNGLPY 613
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 224 bits (571), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 197/670 (29%), Positives = 290/670 (43%), Gaps = 109/670 (16%)
Query 10 IKNKPRRSGFDLGNKNAFTAKVGELLPVYWKFCLPGDKFKISQEWFTRTQPVDTSAFTRI 69
+KNKP R+ FDL ++N FTAKVGELLP + + PGD K+S +FTRT P+ ++AFTR+
Sbjct 13 LKNKPSRNSFDLSHRNMFTAKVGELLPCFVQELNPGDSVKVSSSYFTRTAPLQSNAFTRL 72
Query 70 REYYEWFFVPLHLMYRNSNEAIMSMENQPN------YAAS--GTQSITFNRKLPWVDLLT 121
RE ++FFVP +++ + +++M N A+S G Q +T ++P V+ T
Sbjct 73 RENVQYFFVPYSALWKYFDSQVLNMTKNANGGDISRIASSLVGNQKVT--TQMPCVNYKT 130
Query 122 L--------NNAVENVKASTYHDNMFGFSRALGFAKLYNYLGVGQVDPSKTLANLRI--- 170
L N + S + G R AKL LG G + AN ++
Sbjct 131 LHAYLLKFINRSTVGSDGSVGPEFNRGCYRHAESAKLLQLLGYGNF--PEQFANFKVNND 188
Query 171 --------------------SAFPFYAYQKIYNDYYRNSQWEVNKPWTYNCDFWNGEDTT 210
S F AY KI ND+Y QW+ YN N + T
Sbjct 189 KHNQSGQNFKDVTYNNSPYLSIFRLLAYHKICNDHYLYRQWQ-----PYNASLCNVDYLT 243
Query 211 PVASS----KDLFDTNPNDSI-------FELRYANWNKDLYMGAMPNAQFGDVAFVPVDS 259
P +SS D + P+DSI ++R++N D + G +P +QFG + V ++
Sbjct 244 PNSSSLLSIDDALLSIPDDSIKAEKLNLLDMRFSNLPLDYFTGVLPTSQFGSESVVNLNL 303
Query 260 SGKLPVSLPSIEVGGVAPIYNTGAGGVQPDAQIGLRGAITGAPDNGQTV--TAYGADKTD 317
G A + G D+ G TG + Q V +A G K D
Sbjct 304 G----------NASGSAVL----NGTTSKDS--GRWRTTTGEWEMEQRVASSANGNLKLD 347
Query 318 AARPYFYAVPDGSVAHLKTNAKTIQVPYEFSSKFDVLQLRAAECLQKWKEIAQANGQNYA 377
+ F ++H T + + + S ++ LR A QK+KEI AN ++
Sbjct 348 NSNGTF-------ISHDHTFSGNVAINTSLSGNLSIIALRNALAAQKYKEIQLANDVDFQ 400
Query 378 SQVKAHFGVSPNPMTSHRCQRVCGFDGSIDISAVENTNLSSDEAIIRG---KGIGGYRVN 434
SQV+AHFG+ P+ + + G I+I+ N NLS D G +G G +
Sbjct 401 SQVEAHFGIKPDEKNENSL-FIGGSSSMININEQINQNLSGDNKATYGAAPQGNGSASI- 458
Query 435 KPETFKTTEHGVLMCIYHAVPLLDYAPTGPDLQFMTTVDGDSWPVPELDSVGFEEL---- 490
F +GV++ IY P+LD+A G D T D + +PE+DS+G ++
Sbjct 459 ---KFTAKTYGVVIGIYRCTPVLDFAHLGIDRTLFKT-DASDFVIPEMDSIGMQQTFRCE 514
Query 491 --------PSYSLLNTSDVQPIKEPRPFGYVPRYISWKTSVDVVRGAFIDTLKSWTAPIG 542
+ D +GY PRY +KTS D GAF +LKSW I
Sbjct 515 VAAPAPYNDEFKAFRVGDGSSPDMSETYGYAPRYSEFKTSYDRYNGAFCHSLKSWVTGIN 574
Query 543 EDYMKIYFDNNNVPGGAHFGFYTWFKVNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVA 602
D ++ NN A F P +V +F V + + + DQL V
Sbjct 575 FDAIQ----NNVWNTWAGINAPNMFACRPDIVKNLFLVSSTNNSDDDQLYVGMVNMCYAT 630
Query 603 RNLSYDGLPY 612
RNLS GLPY
Sbjct 631 RNLSRYGLPY 640
>gi|575094339|emb|CDL65730.1| unnamed protein product [uncultured bacterium]
Length=588
Score = 179 bits (454), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 152/548 (28%), Positives = 238/548 (43%), Gaps = 87/548 (16%)
Query 16 RSGFDLGNKNAFTAKVGELLPVYWKFCLPGDKFKISQEWFTRTQPVDTSAFTRIREYYEW 75
++GFD+ ++ FT+ VG+LLPV++ + PGDK +IS FTRTQP+ ++A R+ E+ E+
Sbjct 16 KNGFDMSQRHPFTSSVGQLLPVFYDYLNPGDKIRISANLFTRTQPMKSTAMARLTEHIEY 75
Query 76 FFVPLHLMYRNSNEAIMSMENQPNYAASGTQSITFNRKLPWVDLLTLNNAVE-------- 127
FFVP M+ +++ + + ++T +P+ ++ A+E
Sbjct 76 FFVPFEQMFSLFGSVFYGIDDYNSSSLVKHNNLT----MPFFKSDAVSAALEAAYTSFSS 131
Query 128 NVKASTYHDNMFGFSRALGFAKLYNYLGVGQVDPSKTLA---NLRISAFPFYAYQKIYND 184
++ +M G R G +L LG G + S + +S F F AYQKI+ND
Sbjct 132 SINRKVLTPDMMGQPRVYGILRLSEMLGYGSLLLSNDNNLLPHADMSVFLFTAYQKIFND 191
Query 185 YYRNSQWEVNKPWTYNCDFWNGEDTTPVASSKDLFDTNPNDSIFELRYANWNKDLYMGAM 244
+YR + + +YN D+ G+ T ++S+FEL Y W KD + +
Sbjct 192 FYRLDDYTSVQHKSYNVDYAQGQPIT-------------DNSMFELHYRPWKKDYFTNVI 238
Query 245 PNAQFGDVAFVPVDSSGKLPVSLPSIEVGGVAPIYNTGAGGVQPDAQIGLRGAITGAPDN 304
PN F VD+ GAG D +GL
Sbjct 239 PNPYFSS-----VDNKSSF-----------------GGAGLF--DRPVGL---------- 264
Query 305 GQTVTAYGADKTDAARPYFYAVPDGSVAHLKTNAKTIQ-VPYEFSSK----FDVLQLRAA 359
++T++ D +D F P ++ ++ N Q +P +S V LR
Sbjct 265 --SITSFNFDGSD-----FLQAP-SDLSTMENNQPIFQELPVNLTSASSAGLSVSDLRYL 316
Query 360 ECLQKWKEIAQANGQNYASQVKAHFGVSPNPMTSHRCQRVCGFDGSIDISAVENTNLSSD 419
K I Q G++Y +Q AHFG S + G + IS+VE+T + D
Sbjct 317 YATDKLLRITQFAGKHYDAQTLAHFGKRVPQGVSGEVYYIGGQSQPLQISSVESTATTFD 376
Query 420 EAIIRGKGIG-----GYRV---NKPETFKTTEHGVLMCIYHAVPLLDYAPTGPDLQFMTT 471
+ G +G GY K +F+ HGVLM IY AVP DY D T
Sbjct 377 SGDVVGSVLGELAGKGYSQTGNQKDFSFEAPCHGVLMAIYSAVPEADYLDERIDY-LNTL 435
Query 472 VDGDSWPVPELDSVGFEELPSYSLLNTSDVQPIKEPRPFGYVPRYISWKTSVDVVRGAFI 531
+ + + PE DS+G E P+Y L + + G+ RY K+ D++ GAF
Sbjct 436 IQSNDFYKPEFDSLGMEPFPNYEL---DQYRMVGNNSRLGWRYRYSGLKSKPDLISGAFK 492
Query 532 DTLKSWTA 539
TL+ W A
Sbjct 493 YTLRDWVA 500
>gi|517172762|ref|WP_018361580.1| hypothetical protein [Prevotella nanceiensis]
Length=568
Score = 175 bits (444), Expect = 5e-44, Method: Compositional matrix adjust.
Identities = 171/625 (27%), Positives = 269/625 (43%), Gaps = 105/625 (17%)
Query 16 RSGFDLGNKNAFTAKVGELLPVYWKFCLPGDKFKISQEWFTRTQPVDTSAFTRIREYYEW 75
R+ FD+ ++ FTA G LLPV LP D +I+ F RT P++++AF +R YE+
Sbjct 18 RNAFDISQRHLFTAPAGALLPVLSLDLLPHDHVEINASDFMRTLPMNSAAFMSMRGVYEF 77
Query 76 FFVPLHLMYRNSNEAIMSMENQPN---YAASGT---QSITFN-RKLPWVDLLTLNNAVEN 128
+FVP ++ ++ I M + + YA G ++F+ +KL VD N A ++
Sbjct 78 YFVPYKQLWSGFDQFITGMSDYKSSFMYAFKGKTPPSCVSFDVQKL--VDWCKTNTA-KD 134
Query 129 VKASTYHDNMFGFSRALGFAKLYNYLGVGQVDPSKTLANLRISAFPFYAYQKIYNDYYRN 188
+ + ++ LG+ K N GV +P+ T + + F AYQKIYND+YRN
Sbjct 135 IHGFDKNKGVYRILDLLGYGKYANSAGVPYTNPTSTTMG-KCTPFRGLAYQKIYNDFYRN 193
Query 189 SQWEVNKPWTYNCDFWNGEDTTPVASSKDLFDTNPND----SIFELRYANWNKDLYMGAM 244
+ +E + ++N D + G S + +T PN+ F LRY N KDL
Sbjct 194 TTYEEYQLESFNVDMFYG--------SGKVKETIPNEPWDYDWFTLRYRNAQKDLLTNVR 245
Query 245 PNAQFGDVAFVPVDSSGKLPVSLPSIEVGGVAPIYNTGAGGVQPDAQIGLRGAITGAPDN 304
P P + P + TG +
Sbjct 246 PT---------------------PLFSIDDFNPQFFTGGSDI------------------ 266
Query 305 GQTVTAYGADKTDAARPYFYAVPDGSVAHLKTNAKTIQVPYEFSSKFDVLQLRAAECLQK 364
V G + T Y SV + N K V + + V +R A L+K
Sbjct 267 ---VMEKGPNVTGGTHEY-----RDSVVIVGKNLKENGVDSK-RTMISVADIRNAFALEK 317
Query 365 WKEIAQANGQNYASQVKAHFGVSPNPMTSHRCQRVCGFDGSIDISAVENTNLSSDEAIIR 424
+ G+ Y Q++AHFG+S RC + GFD +I + V ++ ++ +
Sbjct 318 LASVTMRAGKTYKEQMEAHFGISVEEGRDGRCTYIGGFDSNIQVGDVTQSSGTTVTG-TK 376
Query 425 GKGIGGY--RVNKPET--------FKTTEHGVLMCIYHAVPLLDYAPTGPDLQFMTTVDG 474
GGY R T F EHG+LMCIY VP + Y D F+ ++
Sbjct 377 DTSFGGYLGRTTGKATGSGSGHIRFDAKEHGILMCIYSLVPDVQYDSKRVD-PFVQKIER 435
Query 475 DSWPVPELDSVGFEEL----PSYSLLNTSDVQPIKEPRPFGYVPRYISWKTSVDVVRGAF 530
+ VPE +++G + L SY N + IK FG+ PRY +KT++D+ G F
Sbjct 436 GDFFVPEFENLGMQPLFAKNISYKYNNNTANSRIKNLGAFGWQPRYSEYKTALDINHGQF 495
Query 531 I--DTLKSWTA--PIGEDYMKIYFDNNNVPGGAHFGFYTWFKVNPSVVNPIFGVVADGSW 586
+ + L WT GE N N+ + FK+NP ++ +F V +G+
Sbjct 496 VHQEPLSYWTVARARGES-----MSNFNI---------STFKINPKWLDDVFAVNYNGTE 541
Query 587 NTDQLLVNCDFDVRVARNLSYDGLP 611
TDQ+ C F++ ++S DG+P
Sbjct 542 LTDQVFGGCYFNIVKVSDMSIDGMP 566
>gi|647452987|ref|WP_025792807.1| hypothetical protein [Prevotella histicola]
Length=584
Score = 171 bits (433), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 182/659 (28%), Positives = 274/659 (42%), Gaps = 143/659 (22%)
Query 13 KPR--RSGFDLGNKNAFTAKVGELLPVYWKFCLPGDKFKISQEWFTRTQPVDTSAFTRIR 70
KPR R+GFDL ++ F+AK G+LLP+ P + FK S + RT ++T+++ R++
Sbjct 5 KPRLARNGFDLSSRRIFSAKAGQLLPIGCWEVNPSEHFKFSVQDLVRTTTLNTASYARMK 64
Query 71 EYYEWFFVPLHLMYRNSNEAIMSMENQPNYAASGTQ---SITFNRKLPWVDLLTLNNAVE 127
EYY +FFV +++ ++ I+ N P+ A +G + + +N+ V L +
Sbjct 65 EYYHFFFVSYRSLWQWFDQFIVGT-NNPHSALNGVKKNGTTNYNQICSSVPTFDLGKLIT 123
Query 128 NVKASTYHDNMFGFSRALGFAKLYNYLGVGQVDPSK--TLANL----------------- 168
+K S F +S G AKL N L G + K L NL
Sbjct 124 RLKTSDMDSQGFNYSE--GAAKLLNMLNYGVTNKGKFMNLENLITSTSYLPSKDDKEPSS 181
Query 169 ----RISAFPFYAYQKIYNDYYRNSQWEVNKPWTYNCDFWNGEDTTPVASSKDLFDTNPN 224
++S F AYQKI+ND+YRN W + ++N D + + + L
Sbjct 182 IYACKVSPFRLLAYQKIFNDFYRNQDWTPSDVRSFNVDDYADDSNLTIEPDVAL------ 235
Query 225 DSIFELRYANWNKDLYMGAMPNAQFGDVAFVPVDSSGKLPVSLPSIEVG-GVAPIYNTGA 283
++RY + KD P + D F +LP G G + N +
Sbjct 236 -KFCQMRYRPYAKDWLTSMKPTPNYSDGIF-----------NLPEYVRGNGNVILTNNKS 283
Query 284 GGVQPDAQIGLRGAITGAPDNGQTVTAYGADKTDAARPYFYAVPDGSVAHLKTNAKTIQV 343
G V D+ G+V+
Sbjct 284 GSVSLDS--------------------------------------GTVS----------- 294
Query 344 PYEFSSKFDVLQLRAAECLQKWKEIA-QANGQNYASQVKAHFGVSPNPMTSHRCQRVCGF 402
P FS V LRAA L K E +ANG +YASQ++AHFG ++ + + GF
Sbjct 295 PSSFS----VNDLRAAFALDKMLEATRRANGLDYASQIEAHFGFKVPESRANDARFLGGF 350
Query 403 DGSIDISAV--ENTNLSSDEAI-----IRGKGIGGYRVNKPETFKTTEHGVLMCIYHAVP 455
D SI +S V N N +SD + + GKGIG E F +TEHG++MCIY P
Sbjct 351 DNSIVVSEVVSTNGNAASDGSHASIGDLGGKGIGSMSSGTIE-FDSTEHGIIMCIYSVAP 409
Query 456 LLDYAPTGPDLQFMTTVDGDSWPVPELDSVGFEELPSYSLLNTSDVQPIKEP-------- 507
+Y + D F + + + PE +G++ L L+ ++ K+
Sbjct 410 QSEYNASYLD-PFNRKLTREQFYQPEFADLGYQALIGSDLICSTLGMNEKQAGFSDIELN 468
Query 508 -RPFGYVPRYISWKTSVDVVRGAFID--TLKSWTAP-----IGEDYMKIYFDNNNVPGGA 559
GY RY +KT+ D+V G F +L W P G+ KI +N GGA
Sbjct 469 NNLLGYQVRYNEYKTARDLVFGDFESGKSLSYWCTPRFDFGYGDTEKKIAPENK---GGA 525
Query 560 HF----GFYTW----FKVNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGL 610
+ W F +NP++VNPIF A D +VN DV+ R +S GL
Sbjct 526 DYRKKGNRSHWSSRNFYINPNLVNPIFLTSA---VQADHFIVNSFLDVKAVRPMSVTGL 581
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 163 bits (412), Expect = 6e-40, Method: Compositional matrix adjust.
Identities = 165/621 (27%), Positives = 254/621 (41%), Gaps = 111/621 (18%)
Query 16 RSGFDLGNKNAFTAKVGELLPVYWKFCLPGDKFKISQEWFTRTQPVDTSAFTRIREYYEW 75
R+ FDL ++ FTA G LLPV +P D +I+ + F RT P++T+AF +R YE+
Sbjct 17 RNAFDLSQRHLFTAHAGMLLPVLNLDLIPHDHVEINAQDFMRTLPMNTAAFASMRGVYEF 76
Query 76 FFVPLHLMYRNSNEAIMSMENQPNYAASGTQSITFNRKLPWVDLLTLNNAVENVKAS--- 132
FFVP H ++ ++ I M + + A Q T ++P+ ++ ++ N++ K S
Sbjct 77 FFVPYHQLWAQFDQFITGMNDFHSSANKSIQGGTSPLQVPYFNVDSVFNSLNTGKESGSG 136
Query 133 -------TYHDNMFGFSRALGFAKLYNYLGVGQVDPSKTLAN---LRISAFPFYAYQKIY 182
+ F LG+ + ++ G D L N S F AY KIY
Sbjct 137 STDDLQYKFKYGAFRLLDLLGYGRKFDSFGTAYPDNVSGLKNNLDYNCSVFRILAYNKIY 196
Query 183 NDYYRNSQWEVNKPWTYNCDFWNGEDTTPVASSKDLFDTNPNDSIFELRYANWNKDLYMG 242
DYYRNS +E ++N D + G L D +F+LRY N D +
Sbjct 197 QDYYRNSNYENFDTDSFNFDKFKG----------GLVDAKVVADLFKLRYRNAQTDYFTN 246
Query 243 AMPNAQFG-DVAFVPVDSSGKLPVSLPSIEVGGVAPIYNTGAGGVQPDAQIGLRGAITGA 301
+ F AF VD+ +AP R +
Sbjct 247 LRQSQLFSFTTAFEDVDNI-------------NIAP-----------------RDYVKSD 276
Query 302 PDNGQTVTAYGADKTDAARPYFYAVPDGSVAHLKTNAKTIQVPYEFSSKFDVLQLRAAEC 361
N V +G D TD++ D SV+ L+ K + +RA +
Sbjct 277 GSNFTRVN-FGVD-TDSSE------GDFSVSSLRAAFAV--------DKLLSVTMRAGKT 320
Query 362 LQKWKEIAQANGQNYASQVKAHFGVSPNPMTSHRCQRVCGFDGSIDISAVENTNLSSDEA 421
Q Q++AH+GV R + GFD + +S V T+ ++
Sbjct 321 FQ--------------DQMRAHYGVEIPDSRDGRVNYLGGFDSDMQVSDVTQTSGTTATE 366
Query 422 I---------IRGKGIGGYRVNKPETFKTTEHGVLMCIYHAVPLLDYAPTGPDLQFMTTV 472
+ GKG G R F EHGVLMCIY VP + Y T D + +
Sbjct 367 YKPEAGYLGRVAGKGTGSGR--GRIVFDAKEHGVLMCIYSLVPQIQYDCTRLD-PMVDKL 423
Query 473 DGDSWPVPELDSVGFEELPSYSLLNTSDVQPIKEPRPFGYVPRYISWKTSVDVVRGAFI- 531
D + PE +++G + L S + + P K P GY PRY +KT++DV G F
Sbjct 424 DRFDYFTPEFENLGMQPLNSSYISSFCTTDP-KNP-VLGYQPRYSEYKTALDVNHGQFAQ 481
Query 532 -DTLKSWTAPIGEDYMKIYFDNNNVPGGAHFGFYTWFKVNPSVVNPIFGVVADGSWNTDQ 590
D L SW+ + F + FK++P +N IF V +G+ D
Sbjct 482 SDALSSWSVSRFRRWTT--FPQLEIAD---------FKIDPGCLNSIFPVDYNGTEANDC 530
Query 591 LLVNCDFDVRVARNLSYDGLP 611
+ C+F++ ++S DG+P
Sbjct 531 VYGGCNFNIVKVSDMSVDGMP 551
Lambda K H a alpha
0.318 0.136 0.428 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4587133073826