bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-10_CDS_annotation_glimmer3.pl_2_6
Length=164
Score E
Sequences producing significant alignments: (Bits) Value
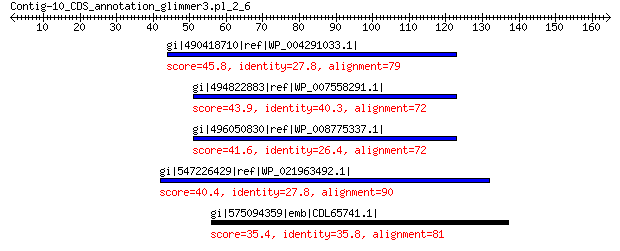
gi|490418710|ref|WP_004291033.1| hypothetical protein 45.8 0.001
gi|494822883|ref|WP_007558291.1| hypothetical protein 43.9 0.006
gi|496050830|ref|WP_008775337.1| hypothetical protein 41.6 0.059
gi|547226429|ref|WP_021963492.1| putative uncharacterized protein 40.4 0.15
gi|575094359|emb|CDL65741.1| unnamed protein product 35.4 9.3
>gi|490418710|ref|WP_004291033.1| hypothetical protein [Bacteroides eggerthii]
gi|217986637|gb|EEC52971.1| hypothetical protein BACEGG_02722 [Bacteroides eggerthii DSM
20697]
Length=155
Score = 45.8 bits (107), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/79 (28%), Positives = 43/79 (54%), Gaps = 0/79 (0%)
Query 44 DTDETRPVRYTSDVCLILHTKDLASRAGLAVASKFGqskqpisqiqqiMDTMSDEDLLAT 103
+ D + +R TSD+ ++ + + L + F ++ + M+DE L +
Sbjct 45 ECDGKKSIRITSDIYMLFNQQRLDKLTRSQLVEYFDNLSVSEPKMSDLRKKMTDEQLCSF 104
Query 104 IRSRYIQSPSEIIAWSKEL 122
++SR+IQ+PSE++AWS+ L
Sbjct 105 VKSRFIQTPSELMAWSQYL 123
>gi|494822883|ref|WP_007558291.1| hypothetical protein [Bacteroides plebeius]
gi|198272098|gb|EDY96367.1| hypothetical protein BACPLE_00803 [Bacteroides plebeius DSM 17135]
Length=140
Score = 43.9 bits (102), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 43/72 (60%), Gaps = 1/72 (1%)
Query 51 VRYTSDVCLILHTKDLASRAGLAVASKFGqskqpisqiqqiMDTMSDEDLLATIRSRYIQ 110
V Y +D+ +IL+ + L S LA S + + SQ+ Q+ + MSDE L ++SRYIQ
Sbjct 31 VTYRNDIYMILNQRRLDS-MTLAQFSDYLDHDRSASQLSQMREKMSDEQLHQFVKSRYIQ 89
Query 111 SPSEIIAWSKEL 122
PSE+ AW+ L
Sbjct 90 HPSELRAWASYL 101
>gi|496050830|ref|WP_008775337.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448894|gb|EEO54685.1| hypothetical protein BSCG_01610 [Bacteroides sp. 2_2_4]
Length=154
Score = 41.6 bits (96), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 40/72 (56%), Gaps = 0/72 (0%)
Query 51 VRYTSDVCLILHTKDLASRAGLAVASKFGqskqpisqiqqiMDTMSDEDLLATIRSRYIQ 110
+R +SD+ ++ + + L + ++ F + ++ + DE L++ ++SR+IQ
Sbjct 51 IRLSSDIYMLFNQQRLDKLSQTSLLEYFNNISVTEPRFNELRSKLGDEQLISFVKSRFIQ 110
Query 111 SPSEIIAWSKEL 122
S SE++AWS L
Sbjct 111 SKSELMAWSNYL 122
>gi|547226429|ref|WP_021963492.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103381|emb|CCY83992.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=152
Score = 40.4 bits (93), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 25/91 (27%), Positives = 46/91 (51%), Gaps = 1/91 (1%)
Query 42 IDDTDETRPVRYTSDVCLILHTKDLASRAGLAVASKFGqskqpisqiqqiMDTMSDEDLL 101
+ +D+ + Y D+ ++ + L S + + ++ + ++DE L+
Sbjct 41 FNGSDKKTAIAYVDDIYMLFNQNRLDSVGRDTIQKWLDGLTPRSDSLAKLRENVTDEQLM 100
Query 102 ATIRSRYIQSPSEIIAWSKELSA-YAENLES 131
+SRYIQS SE++AWS+ L+A YA L S
Sbjct 101 EICKSRYIQSSSELLAWSEYLNANYASILSS 131
>gi|575094359|emb|CDL65741.1| unnamed protein product [uncultured bacterium]
Length=156
Score = 35.4 bits (80), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 29/81 (36%), Positives = 43/81 (53%), Gaps = 7/81 (9%)
Query 56 DVCLILHTKDLASRAGLAVASKFGqskqpisqiqqiMDTMSDEDLLATIRSRYIQSPSEI 115
DV ++ + + L A+A ++ + SQ+ + MSD L A I+SRYIQS SE+
Sbjct 54 DVYMLFNQQRLDRMTNAALADWLSRTSRTDSQLASLRSRMSDVQLGAFIKSRYIQSRSEL 113
Query 116 IAWSKELSAYAENLESQAQEL 136
+ AY+ LESQ EL
Sbjct 114 L-------AYSSYLESQYPEL 127
Lambda K H a alpha
0.312 0.128 0.349 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 432739757718