bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-33_CDS_annotation_glimmer3.pl_2_1
Length=174
Score E
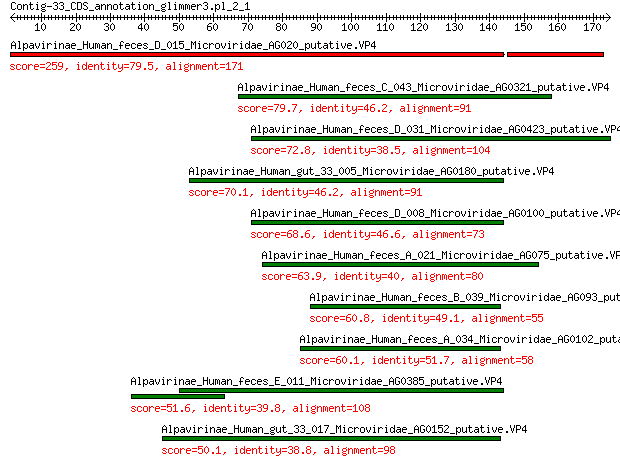
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4 259 2e-85
Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4 79.7 2e-19
Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4 72.8 6e-17
Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4 70.1 4e-16
Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4 68.6 2e-15
Alpavirinae_Human_feces_A_021_Microviridae_AG075_putative.VP4 63.9 6e-14
Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4 60.8 5e-13
Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4 60.1 9e-13
Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4 51.6 6e-10
Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4 50.1 2e-09
> Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4
Length=563
Score = 259 bits (663), Expect = 2e-85, Method: Compositional matrix adjust.
Identities = 126/143 (88%), Positives = 135/143 (94%), Gaps = 0/143 (0%)
Query 1 VSAKVNRPYMLEHLHLIEAERYKALSLRYPNFGSKFRPYILRSILRKSPLQRFKDEYFEE 60
+SA+VNRPYMLEHL L+EA+RYKAL+LRYPNF SK RPYILRSI R S LQ FKDEYFEE
Sbjct 117 MSAEVNRPYMLEHLRLLEADRYKALALRYPNFISKARPYILRSIPRVSKLQNFKDEYFEE 176
Query 61 LVWMLPELAESLKKKNNTDANGAFPQFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKI 120
LVWMLPE+AESLKKKNNTDANGAFPQFKGLLKY+NIRDYQLF+KRLRKYLSKK+GKYEKI
Sbjct 177 LVWMLPEIAESLKKKNNTDANGAFPQFKGLLKYVNIRDYQLFAKRLRKYLSKKVGKYEKI 236
Query 121 HSYVVSEYSPKTLRPHFHILFFF 143
HSYVVSEYSPKT RPHFHILFFF
Sbjct 237 HSYVVSEYSPKTFRPHFHILFFF 259
Score = 22.3 bits (46), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 10/30 (33%), Positives = 14/30 (47%), Gaps = 2/30 (7%)
Query 145 LGRNRP--KHSTGCISELETWSCRYATCEG 172
RN P + + G +SE W C Y +G
Sbjct 397 FARNEPFEEATPGNVSEFICWWCAYNFRQG 426
> Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4
Length=546
Score = 79.7 bits (195), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 42/91 (46%), Positives = 54/91 (59%), Gaps = 4/91 (4%)
Query 67 ELAESLKKKNNTDANGAFPQFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVS 126
+ E+ KK+ N D G +P K + Y++ +D QLF KR+RK + EKIH+Y+V
Sbjct 128 DYIENFKKQANLDVKGCYPHLKDMYGYLSRKDCQLFMKRVRKQIRNYTD--EKIHTYIVG 185
Query 127 EYSPKTLRPHFHILFFFRLGRNRPKHSTGCI 157
EYSPK RPHFHILFFF N S G I
Sbjct 186 EYSPKHFRPHFHILFFF--NSNELSQSFGSI 214
> Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4
Length=532
Score = 72.8 bits (177), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 40/107 (37%), Positives = 56/107 (52%), Gaps = 10/107 (9%)
Query 71 SLKKKNNTDANGAFPQFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSP 130
S +K N NG +P G + Y+ D L+ KR+RKY+SK +G E IH+Y+V EY P
Sbjct 137 SYAQKANLSFNGKYPALSGRIPYLLHGDVSLYMKRVRKYISK-LGINETIHTYIVGEYGP 195
Query 131 KTLRPHFHILFFF---RLGRNRPKHSTGCISELETWSCRYATCEGQR 174
+ RPHFH+L FF L +N + ++ C W C R
Sbjct 196 SSFRPHFHLLLFFDSDELAQNIIRIASSC------WRFGRVDCSASR 236
> Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4
Length=536
Score = 70.1 bits (170), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 42/99 (42%), Positives = 54/99 (55%), Gaps = 10/99 (10%)
Query 53 FKDEYFEELVWMLPELAESLKKKNNTD----ANG----AFPQFKGLLKYINIRDYQLFSK 104
F DE F+ + E +L K + D +G +P LL Y+N RD QLF K
Sbjct 108 FCDEEFDYPTNLRDEAVTALLDKTHLDRTVYPDGRSVVKYPNMGDLLPYLNYRDVQLFHK 167
Query 105 RLRKYLSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFFF 143
R+ + + K EKI+SY V EY PKT RPHFH+LFFF
Sbjct 168 RINQQIKKYTD--EKIYSYTVGEYGPKTFRPHFHLLFFF 204
> Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4
Length=532
Score = 68.6 bits (166), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 34/73 (47%), Positives = 45/73 (62%), Gaps = 1/73 (1%)
Query 71 SLKKKNNTDANGAFPQFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSP 130
S +K N +G +P + Y+ D L+ KR+RKY+SK +G E IH+YVV EY P
Sbjct 137 SYAQKANLSFDGKYPALSDRIPYLLHDDVSLYMKRVRKYISK-LGINETIHTYVVGEYGP 195
Query 131 KTLRPHFHILFFF 143
T RPHFH+L FF
Sbjct 196 VTFRPHFHLLLFF 208
> Alpavirinae_Human_feces_A_021_Microviridae_AG075_putative.VP4
Length=547
Score = 63.9 bits (154), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 32/80 (40%), Positives = 45/80 (56%), Gaps = 4/80 (5%)
Query 74 KKNNTDANGAFPQFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSPKTL 133
+K N ANG +P +K Y++ D LF KRLR KK G+ +HSY+V EY P
Sbjct 145 RKVNLTANGKYPMYKDCFPYLSRYDVALFMKRLRNLFLKKYGQSIYMHSYIVGEYGPVHF 204
Query 134 RPHFHILFFFRLGRNRPKHS 153
RPH+HI+ + N P+ +
Sbjct 205 RPHYHII----ISSNDPRFA 220
> Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4
Length=559
Score = 60.8 bits (146), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 27/55 (49%), Positives = 36/55 (65%), Gaps = 0/55 (0%)
Query 88 KGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFF 142
L+ ++N D Q + KRLRKYL +++GKYE H Y V EY P RPH+H+L F
Sbjct 177 DNLIPFLNYVDVQNYIKRLRKYLFQQLGKYETFHFYAVGEYGPVHFRPHYHLLLF 231
> Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4
Length=539
Score = 60.1 bits (144), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 30/58 (52%), Positives = 37/58 (64%), Gaps = 2/58 (3%)
Query 85 PQFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFF 142
P +GL+ Y+N D QLF KRL + + + EKI+ YVV EY P T RPHFHIL F
Sbjct 149 PYMEGLVGYLNYHDIQLFFKRLNQNIRRITN--EKIYYYVVGEYGPTTFRPHFHILLF 204
> Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4
Length=611
Score = 51.6 bits (122), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 38/102 (37%), Positives = 49/102 (48%), Gaps = 8/102 (8%)
Query 50 LQRFKDEYFEELVWM-LPELAESLKKKNNTDANG-------AFPQFKGLLKYINIRDYQL 101
+ F DE + + M L EL + L K N G + K + + RD +L
Sbjct 152 FRSFDDEPLKFCIPMKLTELQDILIKANGRYDYGKKKVVYPSLADCKLQIPVLQSRDIEL 211
Query 102 FSKRLRKYLSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFFF 143
F KRLR+ L KI YVVSEY P+T RPH+H L FF
Sbjct 212 FFKRLRRNLDSHGFTSSKICYYVVSEYGPQTYRPHWHCLLFF 253
Score = 21.6 bits (44), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 10/27 (37%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 36 FRPYILRSILRKSPLQRFKDEYFEELV 62
F PY+ ++R+S + DE FE +V
Sbjct 74 FLPYLSVEVVRRSGNRYLFDENFETMV 100
> Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4
Length=565
Score = 50.1 bits (118), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 38/99 (38%), Positives = 48/99 (48%), Gaps = 13/99 (13%)
Query 45 LRKSPLQR-FKDEYFEELVWMLPELAESLKKKNNTDANGAFPQFKGLLKYINIRDYQLFS 103
L+ S ++R FKD M P+ S+ K N + P YI RD LF
Sbjct 123 LKNSTVERTFKDPEVRFSYPMKPKELLSILDKINHNVPNRIP-------YICNRDLDLFL 175
Query 104 KRLRKYLSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFF 142
KRLR Y YEK+ Y VSEY P + RPH+H+L F
Sbjct 176 KRLRSYY-----PYEKLRYYAVSEYGPTSYRPHWHLLLF 209
Lambda K H a alpha
0.324 0.139 0.425 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 12344976