bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-29_CDS_annotation_glimmer3.pl_2_5
Length=179
Score E
Sequences producing significant alignments: (Bits) Value
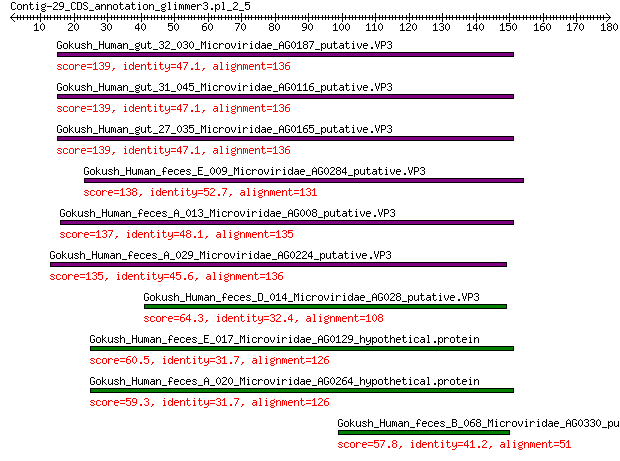
Gokush_Human_gut_32_030_Microviridae_AG0187_putative.VP3 139 4e-43
Gokush_Human_gut_31_045_Microviridae_AG0116_putative.VP3 139 4e-43
Gokush_Human_gut_27_035_Microviridae_AG0165_putative.VP3 139 4e-43
Gokush_Human_feces_E_009_Microviridae_AG0284_putative.VP3 138 5e-43
Gokush_Human_feces_A_013_Microviridae_AG008_putative.VP3 137 2e-42
Gokush_Human_feces_A_029_Microviridae_AG0224_putative.VP3 135 8e-42
Gokush_Human_feces_D_014_Microviridae_AG028_putative.VP3 64.3 4e-15
Gokush_Human_feces_E_017_Microviridae_AG0129_hypothetical.protein 60.5 8e-14
Gokush_Human_feces_A_020_Microviridae_AG0264_hypothetical.protein 59.3 2e-13
Gokush_Human_feces_B_068_Microviridae_AG0330_putative.VP3 57.8 3e-13
> Gokush_Human_gut_32_030_Microviridae_AG0187_putative.VP3
Length=157
Score = 139 bits (349), Expect = 4e-43, Method: Compositional matrix adjust.
Identities = 64/136 (47%), Positives = 91/136 (67%), Gaps = 0/136 (0%)
Query 15 KFANQFEPHARIHTNPGTPEKVLYGPVFDENGTLDLEPKGKENLYDYIQSHKDSCDIKLI 74
+F Q++ RI ++PG+PE + Y +DE G + LE G+ENLYDYIQS+ +SCDI ++
Sbjct 2 EFKTQYDARDRIFSDPGSPEHITYAGHYDEKGRVVLEESGRENLYDYIQSYAESCDIHVL 61
Query 75 VDRCARGDLSALSKAQGMYGDFTTLPRTYAEALQALADAEHFFMRLPVETRAKFDHDPHK 134
+ R A GD+ ALS+ QG YGDF P+TYAEAL + + E FM LPVETR KF + +
Sbjct 62 MKRYANGDVDALSQKQGFYGDFLDFPKTYAEALNHMNEMERQFMALPVETREKFGNSFTE 121
Query 135 FIVSMDKPGFLEKLGV 150
F+ + + F +KLG+
Sbjct 122 FLAASGEADFFDKLGI 137
> Gokush_Human_gut_31_045_Microviridae_AG0116_putative.VP3
Length=157
Score = 139 bits (349), Expect = 4e-43, Method: Compositional matrix adjust.
Identities = 64/136 (47%), Positives = 91/136 (67%), Gaps = 0/136 (0%)
Query 15 KFANQFEPHARIHTNPGTPEKVLYGPVFDENGTLDLEPKGKENLYDYIQSHKDSCDIKLI 74
+F Q++ RI ++PG+PE + Y +DE G + LE G+ENLYDYIQS+ +SCDI ++
Sbjct 2 EFKTQYDARDRIFSDPGSPEHITYAGHYDEKGRVVLEESGRENLYDYIQSYAESCDIHVL 61
Query 75 VDRCARGDLSALSKAQGMYGDFTTLPRTYAEALQALADAEHFFMRLPVETRAKFDHDPHK 134
+ R A GD+ ALS+ QG YGDF P+TYAEAL + + E FM LPVETR KF + +
Sbjct 62 MKRYANGDVDALSQKQGFYGDFLDFPKTYAEALNHMNEMERQFMALPVETREKFGNSFTE 121
Query 135 FIVSMDKPGFLEKLGV 150
F+ + + F +KLG+
Sbjct 122 FLAASGEADFFDKLGI 137
> Gokush_Human_gut_27_035_Microviridae_AG0165_putative.VP3
Length=157
Score = 139 bits (349), Expect = 4e-43, Method: Compositional matrix adjust.
Identities = 64/136 (47%), Positives = 91/136 (67%), Gaps = 0/136 (0%)
Query 15 KFANQFEPHARIHTNPGTPEKVLYGPVFDENGTLDLEPKGKENLYDYIQSHKDSCDIKLI 74
+F Q++ RI ++PG+PE + Y +DE G + LE G+ENLYDYIQS+ +SCDI ++
Sbjct 2 EFKTQYDARDRIFSDPGSPEHITYAGHYDEKGRVVLEESGRENLYDYIQSYAESCDIHVL 61
Query 75 VDRCARGDLSALSKAQGMYGDFTTLPRTYAEALQALADAEHFFMRLPVETRAKFDHDPHK 134
+ R A GD+ ALS+ QG YGDF P+TYAEAL + + E FM LPVETR KF + +
Sbjct 62 MKRYANGDVDALSQKQGFYGDFLDFPKTYAEALNHMNEMERQFMALPVETREKFGNSFTE 121
Query 135 FIVSMDKPGFLEKLGV 150
F+ + + F +KLG+
Sbjct 122 FLAASGEADFFDKLGI 137
> Gokush_Human_feces_E_009_Microviridae_AG0284_putative.VP3
Length=148
Score = 138 bits (348), Expect = 5e-43, Method: Compositional matrix adjust.
Identities = 69/132 (52%), Positives = 84/132 (64%), Gaps = 1/132 (1%)
Query 23 HARIHTNPGTPEKVLYGPVFDENGTLDLEPKGKENLYDYIQSHKDSCDIKLIVDRC-ARG 81
H R + PG E Y P D +G L LE GK N+YD IQSHKDSCDI L++ RC A G
Sbjct 2 HERFPSEPGQREVTTYNPRVDSDGVLHLEESGKINIYDQIQSHKDSCDINLLIQRCVATG 61
Query 82 DLSALSKAQGMYGDFTTLPRTYAEALQALADAEHFFMRLPVETRAKFDHDPHKFIVSMDK 141
D S LS+ QG YGDF+ +P TYA+ L L +A FF LP+ TR KFD + +FI +MDK
Sbjct 62 DESILSRVQGAYGDFSDMPHTYADMLNRLREAREFFDGLPLPTRQKFDCNFEQFISAMDK 121
Query 142 PGFLEKLGVPVP 153
PGFL+ P P
Sbjct 122 PGFLDNFSEPKP 133
> Gokush_Human_feces_A_013_Microviridae_AG008_putative.VP3
Length=160
Score = 137 bits (344), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 65/135 (48%), Positives = 89/135 (66%), Gaps = 0/135 (0%)
Query 16 FANQFEPHARIHTNPGTPEKVLYGPVFDENGTLDLEPKGKENLYDYIQSHKDSCDIKLIV 75
F Q++PH RIH PG E + YG +DE G + L+ G+ + Y IQSH +S D+ +++
Sbjct 2 FRTQYDPHDRIHAEPGQREHIRYGGHYDEKGRVVLDEIGRIDTYAEIQSHAESVDLHVLM 61
Query 76 DRCARGDLSALSKAQGMYGDFTTLPRTYAEALQALADAEHFFMRLPVETRAKFDHDPHKF 135
+R ARGD+ ALSKAQG YGD P+TYAEAL + + E FM LPVE R KF H +F
Sbjct 62 ERYARGDVDALSKAQGFYGDVLDFPKTYAEALNHMNEMERQFMSLPVEIREKFGHSFTEF 121
Query 136 IVSMDKPGFLEKLGV 150
+ S ++P FL+KLG+
Sbjct 122 LASSNEPDFLDKLGI 136
> Gokush_Human_feces_A_029_Microviridae_AG0224_putative.VP3
Length=162
Score = 135 bits (341), Expect = 8e-42, Method: Compositional matrix adjust.
Identities = 62/136 (46%), Positives = 87/136 (64%), Gaps = 0/136 (0%)
Query 13 MPKFANQFEPHARIHTNPGTPEKVLYGPVFDENGTLDLEPKGKENLYDYIQSHKDSCDIK 72
M F NQF H R T G+ EK+ Y ++ G L+L KGKE+ Y YIQSHKDS DI
Sbjct 1 MITFENQFRDHKRFLTGVGSREKITYEARYNAKGQLELNEKGKEDWYGYIQSHKDSVDIH 60
Query 73 LIVDRCARGDLSALSKAQGMYGDFTTLPRTYAEALQALADAEHFFMRLPVETRAKFDHDP 132
++++R RGD+ L++ QG YGD T+ P T+A+AL + +E FF LPVE RAK++H+
Sbjct 61 VLLERFQRGDVDVLNRVQGFYGDITSYPSTFADALNIVRSSEEFFNSLPVEERAKYNHNF 120
Query 133 HKFIVSMDKPGFLEKL 148
+F+ + D P L +L
Sbjct 121 SEFLAAFDSPDTLARL 136
> Gokush_Human_feces_D_014_Microviridae_AG028_putative.VP3
Length=174
Score = 64.3 bits (155), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 35/108 (32%), Positives = 56/108 (52%), Gaps = 0/108 (0%)
Query 41 VFDENGTLDLEPKGKENLYDYIQSHKDSCDIKLIVDRCARGDLSALSKAQGMYGDFTTLP 100
V E G L+ G+EN+Y+ IQ + I+ I+ R GD +AL G+Y D + +P
Sbjct 49 VDSETGAKVLKVVGRENIYEKIQECLEPTKIENIIRRFEEGDPTALGHESGIYADISDMP 108
Query 101 RTYAEALQALADAEHFFMRLPVETRAKFDHDPHKFIVSMDKPGFLEKL 148
EA + + D + F LP++ + KF +DP F+ + L+KL
Sbjct 109 TNIIEAQKRIQDVQAKFASLPIDIKEKFGNDPTVFMAEILSGEGLQKL 156
> Gokush_Human_feces_E_017_Microviridae_AG0129_hypothetical.protein
Length=161
Score = 60.5 bits (145), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 40/131 (31%), Positives = 62/131 (47%), Gaps = 6/131 (5%)
Query 25 RIHTNPGTPEKVLYGPVFDENGTLDLEPKGKENLYDYIQSHKDSCDIKLIVDRCARGDLS 84
R T G ++ + +DE G L + + IQS+ + I+ I++R A D S
Sbjct 13 RKETEAGRKTRLTFRWTYDEKGNKSLVQDEEIDRDAEIQSYLEETKIENIINRAA-FDPS 71
Query 85 ALSKAQGMYGD-----FTTLPRTYAEALQALADAEHFFMRLPVETRAKFDHDPHKFIVSM 139
+ K D FT +P T AEA + AE+ + +LP E + KFD+D KFI
Sbjct 72 IVQKLGAQLSDTEPQDFTNMPSTLAEAQNMMIQAENTWNKLPREIKQKFDNDVEKFIARF 131
Query 140 DKPGFLEKLGV 150
++E LG+
Sbjct 132 GTADWMEALGL 142
> Gokush_Human_feces_A_020_Microviridae_AG0264_hypothetical.protein
Length=157
Score = 59.3 bits (142), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 40/131 (31%), Positives = 61/131 (47%), Gaps = 6/131 (5%)
Query 25 RIHTNPGTPEKVLYGPVFDENGTLDLEPKGKENLYDYIQSHKDSCDIKLIVDRCARGDLS 84
R T G ++ + +DE G L + + IQS+ + I+ I++R A D S
Sbjct 13 RKETEAGRKTRLTFRWTYDEKGNKSLVQDEEIDRDAEIQSYLEETKIENIINRAAY-DPS 71
Query 85 ALSKAQGMYGD-----FTTLPRTYAEALQALADAEHFFMRLPVETRAKFDHDPHKFIVSM 139
+ K D FT +P T AEA + AE + +LP E + KFD+D KFI
Sbjct 72 IVQKLGAQLSDAEPQDFTNMPSTLAEAQNLMIQAEQTWDKLPREVKQKFDNDVDKFIARF 131
Query 140 DKPGFLEKLGV 150
++E LG+
Sbjct 132 GTADWMEALGL 142
> Gokush_Human_feces_B_068_Microviridae_AG0330_putative.VP3
Length=80
Score = 57.8 bits (138), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 38/51 (75%), Gaps = 0/51 (0%)
Query 99 LPRTYAEALQALADAEHFFMRLPVETRAKFDHDPHKFIVSMDKPGFLEKLG 149
+P+TYAE L ++ E+ FM+LPVE RA+F+H +++ +MD+P F++++
Sbjct 1 MPKTYAEVLNSVIAGENAFMKLPVEVRAEFNHSFAEWMAAMDQPNFVDRMA 51
Lambda K H a alpha
0.318 0.136 0.406 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 12901056