bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
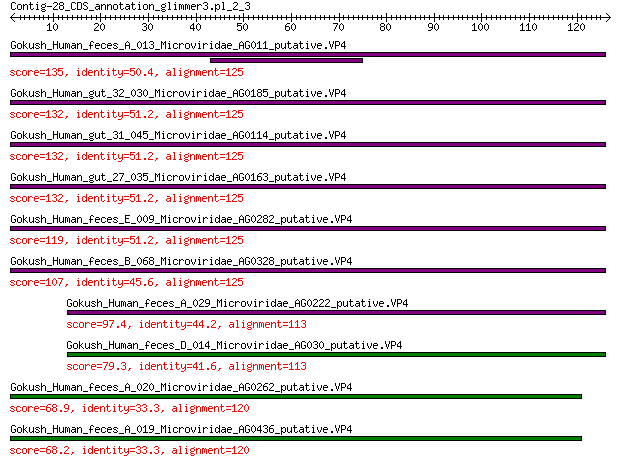
Query= Contig-28_CDS_annotation_glimmer3.pl_2_3
Length=126
Score E
Sequences producing significant alignments: (Bits) Value
Gokush_Human_feces_A_013_Microviridae_AG011_putative.VP4 135 1e-40
Gokush_Human_gut_32_030_Microviridae_AG0185_putative.VP4 132 8e-40
Gokush_Human_gut_31_045_Microviridae_AG0114_putative.VP4 132 8e-40
Gokush_Human_gut_27_035_Microviridae_AG0163_putative.VP4 132 8e-40
Gokush_Human_feces_E_009_Microviridae_AG0282_putative.VP4 119 8e-35
Gokush_Human_feces_B_068_Microviridae_AG0328_putative.VP4 107 2e-30
Gokush_Human_feces_A_029_Microviridae_AG0222_putative.VP4 97.4 9e-27
Gokush_Human_feces_D_014_Microviridae_AG030_putative.VP4 79.3 3e-20
Gokush_Human_feces_A_020_Microviridae_AG0262_putative.VP4 68.9 1e-16
Gokush_Human_feces_A_019_Microviridae_AG0436_putative.VP4 68.2 3e-16
> Gokush_Human_feces_A_013_Microviridae_AG011_putative.VP4
Length=330
Score = 135 bits (339), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 63/125 (50%), Positives = 94/125 (75%), Gaps = 0/125 (0%)
Query 1 VTKKLTGPAACFYDDHNIEPPFTQMSLKPGIGRNYYDNHPDLYKYEYINVSTAKGGRKFR 60
VTKKL FY+ H I P F+ MS +PGI R+YY++HP ++ +YIN+ST KGGRKFR
Sbjct 205 VTKKLNRKEHDFYEKHRICPEFSLMSRRPGIARDYYESHPGVFDSDYINISTPKGGRKFR 264
Query 61 PPKYFDRIFDVEYPDESAAMKEARKRIAMDARQAKLAQTNLSYIDLLAVEERALKERIKS 120
PP+YF+++F++E P S +KE +KR+A+DA+++KL++T+L +LLAVEE+ ++IK
Sbjct 265 PPRYFEKLFEIEDPVRSKELKEIKKRLALDAQKSKLSKTSLELDELLAVEEQNFTDKIKP 324
Query 121 LKRRL 125
L+R L
Sbjct 325 LRRNL 329
Score = 20.0 bits (40), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 16/33 (48%), Gaps = 1/33 (3%)
Query 43 YKYEYINVSTAKGGRKFRP-PKYFDRIFDVEYP 74
YK +++N T K KF P P D+ + P
Sbjct 10 YKSKFVNPETGKAVIKFHPRPDQMDKFEPIALP 42
> Gokush_Human_gut_32_030_Microviridae_AG0185_putative.VP4
Length=310
Score = 132 bits (332), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 64/125 (51%), Positives = 87/125 (70%), Gaps = 0/125 (0%)
Query 1 VTKKLTGPAACFYDDHNIEPPFTQMSLKPGIGRNYYDNHPDLYKYEYINVSTAKGGRKFR 60
V KKL G A FY DHNI+P F+ MS KPGI R Y+D + + +YINVST KGG+KFR
Sbjct 186 VMKKLKGKEAKFYGDHNIQPEFSLMSRKPGIARQYFDENSHCVEEQYINVSTPKGGKKFR 245
Query 61 PPKYFDRIFDVEYPDESAAMKEARKRIAMDARQAKLAQTNLSYIDLLAVEERALKERIKS 120
PP+Y+D++FD+E P++SA +K R ++A A +AKL+ T+L +L VEE R+KS
Sbjct 246 PPRYYDKLFDIECPEKSAELKSLRAKLAQQAMEAKLSNTSLDSYELRDVEEEKQSNRLKS 305
Query 121 LKRRL 125
L+R L
Sbjct 306 LRRNL 310
> Gokush_Human_gut_31_045_Microviridae_AG0114_putative.VP4
Length=310
Score = 132 bits (332), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 64/125 (51%), Positives = 87/125 (70%), Gaps = 0/125 (0%)
Query 1 VTKKLTGPAACFYDDHNIEPPFTQMSLKPGIGRNYYDNHPDLYKYEYINVSTAKGGRKFR 60
V KKL G A FY DHNI+P F+ MS KPGI R Y+D + + +YINVST KGG+KFR
Sbjct 186 VMKKLKGKEAKFYGDHNIQPEFSLMSRKPGIARQYFDENSHCVEEQYINVSTPKGGKKFR 245
Query 61 PPKYFDRIFDVEYPDESAAMKEARKRIAMDARQAKLAQTNLSYIDLLAVEERALKERIKS 120
PP+Y+D++FD+E P++SA +K R ++A A +AKL+ T+L +L VEE R+KS
Sbjct 246 PPRYYDKLFDIECPEKSAELKSLRAKLAQQAMEAKLSNTSLDSYELRDVEEEKQSNRLKS 305
Query 121 LKRRL 125
L+R L
Sbjct 306 LRRNL 310
> Gokush_Human_gut_27_035_Microviridae_AG0163_putative.VP4
Length=310
Score = 132 bits (332), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 64/125 (51%), Positives = 87/125 (70%), Gaps = 0/125 (0%)
Query 1 VTKKLTGPAACFYDDHNIEPPFTQMSLKPGIGRNYYDNHPDLYKYEYINVSTAKGGRKFR 60
V KKL G A FY DHNI+P F+ MS KPGI R Y+D + + +YINVST KGG+KFR
Sbjct 186 VMKKLKGKEAKFYGDHNIQPEFSLMSRKPGIARQYFDENSHCVEEQYINVSTPKGGKKFR 245
Query 61 PPKYFDRIFDVEYPDESAAMKEARKRIAMDARQAKLAQTNLSYIDLLAVEERALKERIKS 120
PP+Y+D++FD+E P++SA +K R ++A A +AKL+ T+L +L VEE R+KS
Sbjct 246 PPRYYDKLFDIECPEKSAELKSLRAKLAQQAMEAKLSNTSLDSYELRDVEEEKQSNRLKS 305
Query 121 LKRRL 125
L+R L
Sbjct 306 LRRNL 310
> Gokush_Human_feces_E_009_Microviridae_AG0282_putative.VP4
Length=340
Score = 119 bits (299), Expect = 8e-35, Method: Compositional matrix adjust.
Identities = 64/125 (51%), Positives = 80/125 (64%), Gaps = 0/125 (0%)
Query 1 VTKKLTGPAACFYDDHNIEPPFTQMSLKPGIGRNYYDNHPDLYKYEYINVSTAKGGRKFR 60
KK G A +Y+ NIEP FT MS KPGIGR Y D HPDLY+Y+ I VST +GG++
Sbjct 216 CMKKADGVDASYYEHFNIEPEFTLMSRKPGIGRMYLDKHPDLYQYQKIFVSTPQGGKEIT 275
Query 61 PPKYFDRIFDVEYPDESAAMKEARKRIAMDARQAKLAQTNLSYIDLLAVEERALKERIKS 120
PKYFDRI E P+ A+KE RK A+ +A + +T+L Y+D L V E K RIKS
Sbjct 276 IPKYFDRIVAQENPEMIEALKEKRKAAAIAKNEAIMKKTDLGYLDYLKVAEDNKKARIKS 335
Query 121 LKRRL 125
L+R L
Sbjct 336 LRRNL 340
> Gokush_Human_feces_B_068_Microviridae_AG0328_putative.VP4
Length=351
Score = 107 bits (268), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 57/125 (46%), Positives = 78/125 (62%), Gaps = 0/125 (0%)
Query 1 VTKKLTGPAACFYDDHNIEPPFTQMSLKPGIGRNYYDNHPDLYKYEYINVSTAKGGRKFR 60
+ KK G A Y+ NIEP F MS KPGI YY++HP+++ Y+ IN+ST GGR FR
Sbjct 227 IMKKQKGQGADVYERFNIEPEFCLMSRKPGIAHQYYEDHPEMWDYDKINISTPNGGRSFR 286
Query 61 PPKYFDRIFDVEYPDESAAMKEARKRIAMDARQAKLAQTNLSYIDLLAVEERALKERIKS 120
PP+YF+R+FDV+ PD S+A K+ + A A + K + SY D++ EE K R K
Sbjct 287 PPQYFERLFDVDCPDLSSARKKKKSEAAKSAEKIKKKLMDKSYSDIMITEENVKKNRTKK 346
Query 121 LKRRL 125
L+R L
Sbjct 347 LRRIL 351
> Gokush_Human_feces_A_029_Microviridae_AG0222_putative.VP4
Length=343
Score = 97.4 bits (241), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 50/113 (44%), Positives = 68/113 (60%), Gaps = 0/113 (0%)
Query 13 YDDHNIEPPFTQMSLKPGIGRNYYDNHPDLYKYEYINVSTAKGGRKFRPPKYFDRIFDVE 72
YD + PFT MS KPGIGR Y+D+HPD Y++INVST GG+KF PP+Y+++++D
Sbjct 231 YDLLGLARPFTLMSRKPGIGRQYFDDHPDCMDYDFINVSTGDGGKKFHPPRYYEKLYDEL 290
Query 73 YPDESAAMKEARKRIAMDARQAKLAQTNLSYIDLLAVEERALKERIKSLKRRL 125
P + K + A A Q K +NL D A+ ER +IKSL+R L
Sbjct 291 EPIAAHERKVKKANAARHAEQIKQKHSNLDEYDRRALAERIKAGQIKSLRRTL 343
> Gokush_Human_feces_D_014_Microviridae_AG030_putative.VP4
Length=348
Score = 79.3 bits (194), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 47/116 (41%), Positives = 70/116 (60%), Gaps = 3/116 (3%)
Query 13 YDDHNIEPPFTQMSLKPGIGRNYYDNHPD-LYKYEYINVSTAKGGRKFR-PPKYFDRIFD 70
Y + I P F S KPGIG YY+ H D +Y + I + AKGG + R PPKYFDR+F
Sbjct 233 YAEAGINPEFCLCSRKPGIGYGYYEAHKDEIYSKDGIAYAKAKGGAQTRKPPKYFDRLFK 292
Query 71 VEYPDESAAMKEARKRIAMDARQAKL-AQTNLSYIDLLAVEERALKERIKSLKRRL 125
+E PD+ A ++ RK +A + +L +T L I+ +EE+ ++ IK+L+R+L
Sbjct 293 LENPDKFAEIQALRKEVAEHQFKYRLVGKTTLPRIEYYKLEEQVKQDTIKALQRKL 348
> Gokush_Human_feces_A_020_Microviridae_AG0262_putative.VP4
Length=338
Score = 68.9 bits (167), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 40/121 (33%), Positives = 68/121 (56%), Gaps = 5/121 (4%)
Query 1 VTKKLTGPAACFYDDHNIEPPFTQMSLKPGIGRNYY-DNHPDLYKYEYINVSTAKGGRKF 59
VTKK+ G + + PPF MSLKPG+G YY N ++K YI +S G++
Sbjct 218 VTKKI-GEETMEHIKRGLRPPFAMMSLKPGLGEEYYLQNKEQIWKQGYIQLSN---GKRA 273
Query 60 RPPKYFDRIFDVEYPDESAAMKEARKRIAMDARQAKLAQTNLSYIDLLAVEERALKERIK 119
P+YF++ + E P+ +K R++ +MD+ ++K+ QT++ L+ +ER + + K
Sbjct 274 AIPRYFEKQMEAEDPERLWEIKRQRQQKSMDSTKSKMEQTDIKLESYLSAQERKIHKFRK 333
Query 120 S 120
S
Sbjct 334 S 334
> Gokush_Human_feces_A_019_Microviridae_AG0436_putative.VP4
Length=338
Score = 68.2 bits (165), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 40/121 (33%), Positives = 69/121 (57%), Gaps = 5/121 (4%)
Query 1 VTKKLTGPAACFYDDHNIEPPFTQMSLKPGIGRNYY-DNHPDLYKYEYINVSTAKGGRKF 59
VTKK+ G + + PPF MSLKPG+G YY N +++ YI +S G++
Sbjct 218 VTKKI-GDETMEHIKRGLRPPFAMMSLKPGLGEEYYLQNKERIWEQGYIQLSN---GKRA 273
Query 60 RPPKYFDRIFDVEYPDESAAMKEARKRIAMDARQAKLAQTNLSYIDLLAVEERALKERIK 119
P+YF++ + E P++ +K R++ AMD+ + K+ QT++ L+ +ER +++ K
Sbjct 274 AIPRYFEKQMEAENPEKLWEIKRQRQQKAMDSTKNKMEQTDIKLEGYLSAQERKIRKFRK 333
Query 120 S 120
S
Sbjct 334 S 334
Lambda K H a alpha
0.319 0.136 0.397 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 7605572