bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
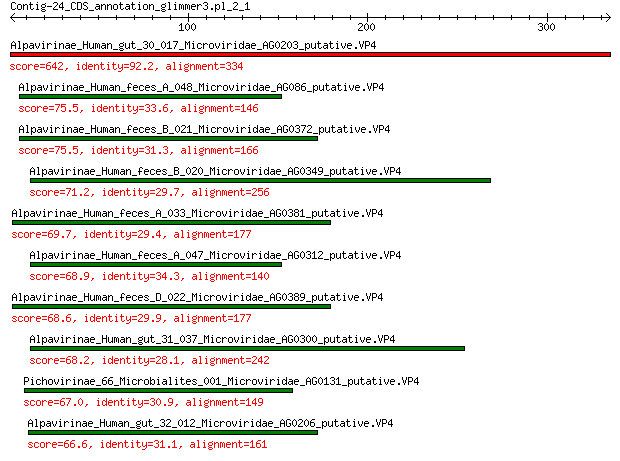
Query= Contig-24_CDS_annotation_glimmer3.pl_2_1
Length=334
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_gut_30_017_Microviridae_AG0203_putative.VP4 642 0.0
Alpavirinae_Human_feces_A_048_Microviridae_AG086_putative.VP4 75.5 3e-17
Alpavirinae_Human_feces_B_021_Microviridae_AG0372_putative.VP4 75.5 4e-17
Alpavirinae_Human_feces_B_020_Microviridae_AG0349_putative.VP4 71.2 9e-16
Alpavirinae_Human_feces_A_033_Microviridae_AG0381_putative.VP4 69.7 2e-15
Alpavirinae_Human_feces_A_047_Microviridae_AG0312_putative.VP4 68.9 5e-15
Alpavirinae_Human_feces_D_022_Microviridae_AG0389_putative.VP4 68.6 7e-15
Alpavirinae_Human_gut_31_037_Microviridae_AG0300_putative.VP4 68.2 8e-15
Pichovirinae_66_Microbialites_001_Microviridae_AG0131_putative.VP4 67.0 2e-14
Alpavirinae_Human_gut_32_012_Microviridae_AG0206_putative.VP4 66.6 3e-14
> Alpavirinae_Human_gut_30_017_Microviridae_AG0203_putative.VP4
Length=363
Score = 642 bits (1655), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 308/336 (92%), Positives = 319/336 (95%), Gaps = 2/336 (1%)
Query 1 MNRPWDYFTQRIMVPCGRCEECLRQQRNDWYVRLERETKYQKSLHRNSVFVTITIAPEYY 60
MNRPWDYFTQRIMVPCG CEECLRQQRNDWY+RLERETKY KSLH NSVFVTITIAPEYY
Sbjct 28 MNRPWDYFTQRIMVPCGHCEECLRQQRNDWYIRLERETKYHKSLHNNSVFVTITIAPEYY 87
Query 61 DSALLNPSSFIRMWFERIRRCFGHSIKHAVFQEFGMHPEQGNEPRLHFHGVLWDVSYSYN 120
DSAL +PSSFIRMWFERIRR FGHSIKHAVFQEFGMHPEQG EPRLHFHG+LWDVSYSYN
Sbjct 88 DSALQDPSSFIRMWFERIRRRFGHSIKHAVFQEFGMHPEQGREPRLHFHGILWDVSYSYN 147
Query 121 AIREAVKDLGFVWISSITDKRLRYVVKYVGKSVYMDDSSAVFAKSLPITVGKLNTNLYDF 180
AIREAVKDLGFVWI+S+TDKRLRYVVKYVGKSVYMD+SSA FAKSLPIT GKLNTNLYDF
Sbjct 148 AIREAVKDLGFVWIASVTDKRLRYVVKYVGKSVYMDESSAAFAKSLPITAGKLNTNLYDF 207
Query 181 LQNRKYRRKFVSPGVGDYLGDFKAPGVTSGLWSYTDYRTGVVYRYRIPRYYDKYLSQDAL 240
LQNRKY RKFVSPGVGDYLGDFKAPGVTSGLWSYTDY+TGVVYRYRIPRYYDKYLSQDAL
Sbjct 208 LQNRKYHRKFVSPGVGDYLGDFKAPGVTSGLWSYTDYKTGVVYRYRIPRYYDKYLSQDAL 267
Query 241 FFRKISTAWTYACVFGSSLALGFLREVAQRVLRPSSFSRIVKGGFSRLVKLREFLS--KV 298
FFRKISTAWTYA FGSSLALGFLREVA RVL PSSFSR+VKGGFSRLVKLR+FLS KV
Sbjct 268 FFRKISTAWTYASAFGSSLALGFLREVAARVLSPSSFSRVVKGGFSRLVKLRDFLSKVKV 327
Query 299 KFSQRFIAVVPDVIDFWVDCFGVNSSNPFFNKIVYG 334
K SQRFIAV PDVIDFWVDCFG++SSNPFFNKIVYG
Sbjct 328 KVSQRFIAVTPDVIDFWVDCFGIDSSNPFFNKIVYG 363
> Alpavirinae_Human_feces_A_048_Microviridae_AG086_putative.VP4
Length=332
Score = 75.5 bits (184), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 49/157 (31%), Positives = 78/157 (50%), Gaps = 22/157 (14%)
Query 6 DYFTQRIMVPCGRCEECLRQQRNDWYVRLERETKYQKSLHRNSVFVTITIAPEYYDSALL 65
D T I CG+C EC +Q++ +W VR+ E + + N+ F+T+TI+ E Y+ L
Sbjct 28 DPRTAYITAACGKCLECRKQKQREWLVRMSEELRTEP----NAYFMTLTISDENYE-ILK 82
Query 66 N----------PSSFIRMWFERIRRCFGHSIKHAVFQEFGMHPEQGNEPRLHFHGVLWDV 115
N + IR+ ERIR+ G SIKH E G + RLH HG++W +
Sbjct 83 NICKSEDDNTIATKAIRLMLERIRKKTGKSIKHWFITELGHEKTE----RLHLHGIVWGI 138
Query 116 SYSYNAIREAVKDLGFVWISS-ITDKRLRYVVKYVGK 151
+ + E + G + + + +K + YV KY+ K
Sbjct 139 --GTDQLIEEKWNYGITYTGNYVNEKTINYVTKYMTK 173
> Alpavirinae_Human_feces_B_021_Microviridae_AG0372_putative.VP4
Length=332
Score = 75.5 bits (184), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 52/177 (29%), Positives = 86/177 (49%), Gaps = 25/177 (14%)
Query 6 DYFTQRIMVPCGRCEECLRQQRNDWYVRLERETKYQKSLHRNSVFVTITIAPEYYDSALL 65
D T I CG+C EC +Q++ +W VR+ E + + N+ F+T+TI+ E Y+ L
Sbjct 28 DPRTAYITAACGKCLECRKQKQREWLVRMSEELRTEP----NAYFMTLTISDENYE-ILK 82
Query 66 N----------PSSFIRMWFERIRRCFGHSIKHAVFQEFGMHPEQGNEPRLHFHGVLWDV 115
N + IR+ ERIR+ G SI+H E G + RLH HG++W +
Sbjct 83 NICKSEDENTIATKAIRLMLERIRKKIGKSIRHWFITELGHEKTE----RLHLHGIVWGI 138
Query 116 SYSYNAIREAVKDLGFVWISS-ITDKRLRYVVKYVGKSVYMDDSSAVFAKSLPITVG 171
+ IRE + G + + + +K + Y+ KY+ K +D+ F + + G
Sbjct 139 G-TDQLIREKW-NYGITYTGNFVNEKTINYITKYMTK---IDEEHPNFVGKVLCSKG 190
> Alpavirinae_Human_feces_B_020_Microviridae_AG0349_putative.VP4
Length=332
Score = 71.2 bits (173), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 76/283 (27%), Positives = 116/283 (41%), Gaps = 69/283 (24%)
Query 12 IMVPCGRCEECLRQQRNDWYVRLERETKYQKSLHRNSVFVTITIAPEYYD--SALLN--- 66
+ CG+C EC +Q++ W VR+ E + N+ F+T+TI E Y+ + + N
Sbjct 34 VTAACGKCMECRQQKQRQWLVRMSEELRQNP----NAYFMTLTIDDENYNKLANICNSKD 89
Query 67 ----PSSFIRMWFERIRRCFGHSIKHAVFQEFGMHPEQGNEPRLHFHGVLWDVSYSYNAI 122
+ +R+ ERIR+ G SIKH E G + RLH HG++W + + I
Sbjct 90 NNEIATKAVRLMLERIRKKTGKSIKHWFITELGHEKTE----RLHLHGIVWGIG-TDQLI 144
Query 123 REAVKDLGFVWISS-ITDKRLRYVVKYVGKSVYMDDSSAVFAKSLPITVGKLNTNLYDFL 181
E + GFV+ + + + + Y+ KY+ K V +D P VG++
Sbjct 145 SEKW-NYGFVYTGNFVNEATINYITKYMTK-VDIDH---------PDFVGQV-------- 185
Query 182 QNRKYRRKFVSPGVGDYLGDFKAPGVTSGLWSYTDYRTGVVYRYR------IPRYYDKYL 235
S G+G G K + YT +T YR R +P YY L
Sbjct 186 --------LCSKGIG--AGYTKREDANNH--KYTKGKTNETYRLRNGAKINLPIYYRNQL 233
Query 236 ----SQDALFFRKISTAWTYACVFGSSLA-------LGFLREV 267
++ LF KI Y V G + LG L E
Sbjct 234 FSEEEREMLFLDKIEKGIIY--VMGQKVHRDNEAEYLGLLEEA 274
> Alpavirinae_Human_feces_A_033_Microviridae_AG0381_putative.VP4
Length=307
Score = 69.7 bits (169), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 52/198 (26%), Positives = 88/198 (44%), Gaps = 31/198 (16%)
Query 2 NRPWDYFTQRIMVPCGRCEECLRQQRNDWYVRLERETKYQKSLHRNSVFVTITIAPEYYD 61
N+P DY ++ VPCG+C +C+++++ W+VR K N+ F T T+ PE+Y+
Sbjct 29 NQP-DY---KLQVPCGKCVQCIKKRQQHWFVRAHNIYKRLGYNLSNTYFCTFTLKPEFYE 84
Query 62 SALLNPSSFIRMWFERIRRCFGHSIKHAVFQEF-----------------GMHPEQ---G 101
+ P +FIR + +R+R+ + F G Q
Sbjct 85 AFCKEPYAFIRRFIDRMRKDQSLRYRDPDTGRFRYRKLSFPYLFVLEVADGKRAAQRKLS 144
Query 102 NEPRLHFHGVLWDVSYSYNAIREAVKDLGFVWISSITD-KRLRYVVKYVGKSVYMDDSSA 160
+E RLH H +++ + ++R G W+S + +RY +KYV K SA
Sbjct 145 SEHRLHIHAIMFGCPLPWWSVRHYWMSFGLAWVSRLRHFGGVRYAMKYVTK------KSA 198
Query 161 VFAKSLPITVGKLNTNLY 178
V +P + L+ LY
Sbjct 199 VHRNDVPNDILDLHGRLY 216
> Alpavirinae_Human_feces_A_047_Microviridae_AG0312_putative.VP4
Length=332
Score = 68.9 bits (167), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 48/157 (31%), Positives = 72/157 (46%), Gaps = 34/157 (22%)
Query 12 IMVPCGRCEECLRQQRNDWYVRLERETKYQKSLHRNSVFVTITIAPEYYDSALLN----- 66
I CG+C EC +Q++ W VR+ E + + N+ F+T+TI + Y S L N
Sbjct 34 ITAACGKCMECRQQKQRQWLVRMSEELRQEP----NAYFITLTIDDKSY-SELSNTYNIT 88
Query 67 -----PSSFIRMWFERIRRCFGHSIKHAVFQEFGMHPEQGNEPRLHFHGVLWDVSYSYNA 121
+ IR+ ERIR+ G SIKH E G + RLH HG++W +
Sbjct 89 DNNEIATKAIRLCLERIRKQTGKSIKHWFITELGHEKTE----RLHLHGIVWGIGTD--- 141
Query 122 IREAVKDLGFVWISSIT-------DKRLRYVVKYVGK 151
K + W IT +K ++Y+ KY+ K
Sbjct 142 -----KLITSKWNYGITFTGFFVNEKTIQYITKYMTK 173
> Alpavirinae_Human_feces_D_022_Microviridae_AG0389_putative.VP4
Length=307
Score = 68.6 bits (166), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 53/201 (26%), Positives = 90/201 (45%), Gaps = 37/201 (18%)
Query 2 NRPWDYFTQRIMVPCGRCEECLRQQRNDWYVRLERETKYQKSLHRNSVFVTITIAPEYYD 61
N+P DY ++ VPCG+C +C+++++ W+VR K N+ F T T+ PE+Y+
Sbjct 29 NQP-DY---KLQVPCGKCVQCIKKRQQHWFVRAHNIYKRLGYNLSNTYFCTFTLKPEFYE 84
Query 62 SALLNPSSFIRMWFERIRR--------------CFGHSIKHAVFQEFGMHPEQG------ 101
+ P +FIR + +R+R+ C+ I F F + G
Sbjct 85 AFCKEPYAFIRRFIDRMRKDQLLRYRNPDTGRFCY-RKISFPYF--FVLEVADGKRAAQR 141
Query 102 ---NEPRLHFHGVLWDVSYSYNAIREAVKDLGFVWISSITD-KRLRYVVKYVGKSVYMDD 157
+E RLH H +++ + +R G W++ + +RY +KY+ K
Sbjct 142 RLHSEHRLHLHAIMFGCPLPWWRVRHYWMSFGLAWVNPLRHFGAVRYAMKYITK------ 195
Query 158 SSAVFAKSLPITVGKLNTNLY 178
SAV +P V L+ LY
Sbjct 196 KSAVHWNDVPKEVLDLHGRLY 216
> Alpavirinae_Human_gut_31_037_Microviridae_AG0300_putative.VP4
Length=304
Score = 68.2 bits (165), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 68/263 (26%), Positives = 110/263 (42%), Gaps = 62/263 (24%)
Query 12 IMVPCGRCEECLRQQRNDWYVRLERETKYQKSLHRNSVFVTITIAPEYYD---------S 62
+ CG+C EC +Q+ W VRL E + N++FVT+TI+ E ++ S
Sbjct 34 VTAACGKCYECRKQKGRAWQVRLSEEVRSDP----NAIFVTLTISDESWEKIKNTYIQLS 89
Query 63 ALLNPSSFIRMWFERIRRCFGHSIKHAVFQEFGMHPEQGNEPRLHFHGVLWDVSYSYNAI 122
+R++ ER+R+ S+KH + E G N R H HG++W + A+
Sbjct 90 DEDCIKKMVRLFLERVRKKTKKSLKHWLTTERG----GTNTERYHLHGLIW--GENTEAL 143
Query 123 REAVKDLGFVWISS-ITDKRLRYVVKYVGKSVYMDDSSAVFAKSLPITVGKLNTNLYDFL 181
+++ GFV+I + + + + Y+ KY+ K+ D F PIT+
Sbjct 144 TKSLWQYGFVFIGTFVNECTVNYITKYITKT---DKKHKDFE---PITL----------- 186
Query 182 QNRKYRRKFVSPGVGD-YLGDFKAPGVTSGLWSYTDYRTGVVYRYR------IPRYYDKY 234
S G+G YL S L + + +T YR R +P YY
Sbjct 187 ---------CSAGIGKGYLSR-----SDSELNRFREGKTTETYRLRNGTKLNLPIYYRNK 232
Query 235 L----SQDALFFRKISTAWTYAC 253
L ++ LF KI + C
Sbjct 233 LYTDEEREKLFLEKIKKGKVWIC 255
> Pichovirinae_66_Microbialites_001_Microviridae_AG0131_putative.VP4
Length=291
Score = 67.0 bits (162), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 46/153 (30%), Positives = 78/153 (51%), Gaps = 15/153 (10%)
Query 9 TQRIMVPCGRCEECLRQQRNDWYVRLERETKYQKSLHRNSVFVTITI----APEYYDSAL 64
T+++ VPCGRC C ++ NDW RL + +K + NS FVT+T P + +
Sbjct 30 TEKVPVPCGRCPNCKLRRVNDWVFRLMQHSK----VSDNSHFVTLTYDTRHVPITENGFM 85
Query 65 LNPSSFIRMWFERIRRCFGHSIKHAVFQEFGMHPEQGNEPRLHFHGVLWDVSYSYNAIRE 124
+ ++ +R+R+ +K+ E+G N R H+H +L+ VS N I +
Sbjct 86 TLDKDAVPLFMKRLRKYETAQLKYYAVGEYGT-----NNKRPHYHMILFGVSDIEN-IHK 139
Query 125 AVKDLGFVWISSITDKRLRYVVKYVGKSVYMDD 157
A + G VW+ ++T + Y +KY+ KS + D
Sbjct 140 AW-NYGSVWVGTVTGDSIAYTMKYIDKSRWRPD 171
> Alpavirinae_Human_gut_32_012_Microviridae_AG0206_putative.VP4
Length=306
Score = 66.6 bits (161), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 50/193 (26%), Positives = 87/193 (45%), Gaps = 39/193 (20%)
Query 11 RIMVPCGRCEECLRQQRNDWYVRLERETKYQKSLHRNSVFVTITIAPEYYDSALLNPSSF 70
++ VPCG C+ECL++++ W++R + K K +F T +I PE Y+ P
Sbjct 34 KLRVPCGHCDECLKKRQQQWFLRADHVVKRLKLRPDQCLFCTFSIKPEVYEQTKERPYLA 93
Query 71 IRMWFERIRRCFGHSIKHAVFQEFGMHP------------------------EQGNEP-- 104
IR + +R+R KH F+E G E+G E
Sbjct 94 IRRFMDRLR-------KHPRFREKGASGRYRYRKVRFPYIFVVEFADGKRARERGLESTH 146
Query 105 RLHFHGVLWDVSYSYNAIREAVKD-LGFVWISSI-TDKRLRYVVKYVGKSV----YMDDS 158
R+HFH +L++ + +R+ + +G W+ + ++ +RYV+KY+ K Y+ D
Sbjct 147 RMHFHAILFNCPLYWWQVRDLWESCVGRAWVDPLHSEAGIRYVLKYMTKDCKAHQYISDI 206
Query 159 SAVFAKSLPITVG 171
A L ++ G
Sbjct 207 DARKNGKLFVSHG 219
Lambda K H a alpha
0.327 0.141 0.450 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 28557990