bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-22_CDS_annotation_glimmer3.pl_2_6
Length=345
Score E
Sequences producing significant alignments: (Bits) Value
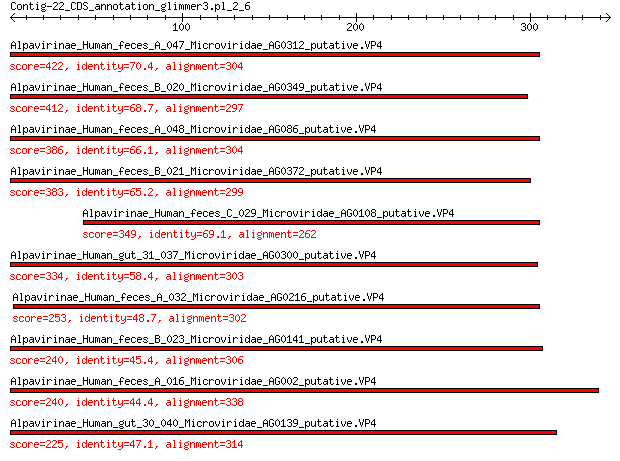
Alpavirinae_Human_feces_A_047_Microviridae_AG0312_putative.VP4 422 4e-149
Alpavirinae_Human_feces_B_020_Microviridae_AG0349_putative.VP4 412 5e-145
Alpavirinae_Human_feces_A_048_Microviridae_AG086_putative.VP4 386 6e-135
Alpavirinae_Human_feces_B_021_Microviridae_AG0372_putative.VP4 383 1e-133
Alpavirinae_Human_feces_C_029_Microviridae_AG0108_putative.VP4 349 7e-121
Alpavirinae_Human_gut_31_037_Microviridae_AG0300_putative.VP4 334 9e-115
Alpavirinae_Human_feces_A_032_Microviridae_AG0216_putative.VP4 253 6e-83
Alpavirinae_Human_feces_B_023_Microviridae_AG0141_putative.VP4 240 4e-78
Alpavirinae_Human_feces_A_016_Microviridae_AG002_putative.VP4 240 2e-77
Alpavirinae_Human_gut_30_040_Microviridae_AG0139_putative.VP4 225 2e-71
> Alpavirinae_Human_feces_A_047_Microviridae_AG0312_putative.VP4
Length=332
Score = 422 bits (1086), Expect = 4e-149, Method: Compositional matrix adjust.
Identities = 214/305 (70%), Positives = 256/305 (84%), Gaps = 3/305 (1%)
Query 1 MCLYPKLIPNKRYLPTKKNGGVPPVCPDERLRYVTAACGDCYECRKQKQRQWVVRMSEEN 60
MCLYPKLI N+RY+P KKNGGVPP CPDERLRY+TAACG C ECR+QKQRQW+VRMSEE
Sbjct 1 MCLYPKLIKNRRYVPNKKNGGVPPQCPDERLRYITAACGKCMECRQQKQRQWLVRMSEEL 60
Query 61 RQTPNAYFLTLTIddksykqlkqkyklkdnndIATKAIRLCLERVRKLTGKSVKHWFITE 120
RQ PNAYF+TLTIDDKSY +L Y + DNN+IATKAIRLCLER+RK TGKS+KHWFITE
Sbjct 61 RQEPNAYFITLTIDDKSYSELSNTYNITDNNEIATKAIRLCLERIRKQTGKSIKHWFITE 120
Query 121 LGHEKTERLHLHGIVWGLGNGEKITNNWKYGITFTGYFVNEKTINYITKYMLKIDEKHPK 180
LGHEKTERLHLHGIVWG+G + IT+ W YGITFTG+FVNEKTI YITKYM KIDE+H
Sbjct 121 LGHEKTERLHLHGIVWGIGTDKLITSKWNYGITFTGFFVNEKTIQYITKYMTKIDEQHKD 180
Query 181 FRGKVLCSAGIGSGYLKREDAKRHVYIPGKTNESYRMKNGGKLNLPIYYRNKIFTEEERE 240
F GKVLCS GIG+GY+KR+DAK+H Y G+T E+YR++NG K+NLPIYYRN++FTEEERE
Sbjct 181 FIGKVLCSKGIGAGYIKRDDAKKHTYKRGETIETYRLRNGSKINLPIYYRNQLFTEEERE 240
Query 241 KLFLDKIEKGIVYVLGSKIDLKTEESRYMGVLLSERERCE-RLYHDSPKDWDKRKYLNRL 299
LFLDKIEKGI+YV+G K+ EE Y LL E + E RLY + ++W+++KYL+RL
Sbjct 241 ALFLDKIEKGIIYVMGQKVHRDDEE--YYLQLLDEGRKTECRLYGYNLQNWEQQKYLSRL 298
Query 300 KKQRQ 304
++Q++
Sbjct 299 RRQKK 303
> Alpavirinae_Human_feces_B_020_Microviridae_AG0349_putative.VP4
Length=332
Score = 412 bits (1059), Expect = 5e-145, Method: Compositional matrix adjust.
Identities = 204/297 (69%), Positives = 242/297 (81%), Gaps = 1/297 (0%)
Query 1 MCLYPKLIPNKRYLPTKKNGGVPPVCPDERLRYVTAACGDCYECRKQKQRQWVVRMSEEN 60
MCLYPKLI NKRYLP KKNGGVPPVCPDERL YVTAACG C ECR+QKQRQW+VRMSEE
Sbjct 1 MCLYPKLIRNKRYLPNKKNGGVPPVCPDERLLYVTAACGKCMECRQQKQRQWLVRMSEEL 60
Query 61 RQTPNAYFLTLTIddksykqlkqkyklkdnndIATKAIRLCLERVRKLTGKSVKHWFITE 120
RQ PNAYF+TLTIDD++Y +L KDNN+IATKA+RL LER+RK TGKS+KHWFITE
Sbjct 61 RQNPNAYFMTLTIDDENYNKLANICNSKDNNEIATKAVRLMLERIRKKTGKSIKHWFITE 120
Query 121 LGHEKTERLHLHGIVWGLGNGEKITNNWKYGITFTGYFVNEKTINYITKYMLKIDEKHPK 180
LGHEKTERLHLHGIVWG+G + I+ W YG +TG FVNE TINYITKYM K+D HP
Sbjct 121 LGHEKTERLHLHGIVWGIGTDQLISEKWNYGFVYTGNFVNEATINYITKYMTKVDIDHPD 180
Query 181 FRGKVLCSAGIGSGYLKREDAKRHVYIPGKTNESYRMKNGGKLNLPIYYRNKIFTEEERE 240
F G+VLCS GIG+GY KREDA H Y GKTNE+YR++NG K+NLPIYYRN++F+EEERE
Sbjct 181 FVGQVLCSKGIGAGYTKREDANNHKYTKGKTNETYRLRNGAKINLPIYYRNQLFSEEERE 240
Query 241 KLFLDKIEKGIVYVLGSKIDLKTEESRYMGVLLSERERCERLYHDSPKDWDKRKYLN 297
LFLDKIEKGI+YV+G K+ + E+ Y+G+L R+ +RLY ++W+++KYLN
Sbjct 241 MLFLDKIEKGIIYVMGQKVH-RDNEAEYLGLLEEARKTEQRLYGVHEQEWEEQKYLN 296
> Alpavirinae_Human_feces_A_048_Microviridae_AG086_putative.VP4
Length=332
Score = 386 bits (992), Expect = 6e-135, Method: Compositional matrix adjust.
Identities = 201/305 (66%), Positives = 242/305 (79%), Gaps = 3/305 (1%)
Query 1 MCLYPKLIPNKRYLPTKKNGGVPPVCPDERLRYVTAACGDCYECRKQKQRQWVVRMSEEN 60
MCLYPKLI NK+YLPTKKN PP D R Y+TAACG C ECRKQKQR+W+VRMSEE
Sbjct 1 MCLYPKLIRNKKYLPTKKNNYNPPKMVDPRTAYITAACGKCLECRKQKQREWLVRMSEEL 60
Query 61 RQTPNAYFLTLTIddksykqlkqkyklkdnndIATKAIRLCLERVRKLTGKSVKHWFITE 120
R PNAYF+TLTI D++Y+ LK K +D+N IATKAIRL LER+RK TGKS+KHWFITE
Sbjct 61 RTEPNAYFMTLTISDENYEILKNICKSEDDNTIATKAIRLMLERIRKKTGKSIKHWFITE 120
Query 121 LGHEKTERLHLHGIVWGLGNGEKITNNWKYGITFTGYFVNEKTINYITKYMLKIDEKHPK 180
LGHEKTERLHLHGIVWG+G + I W YGIT+TG +VNEKTINY+TKYM KIDEKHP
Sbjct 121 LGHEKTERLHLHGIVWGIGTDQLIEEKWNYGITYTGNYVNEKTINYVTKYMTKIDEKHPD 180
Query 181 FRGKVLCSAGIGSGYLKREDAKRHVYIPGKTNESYRMKNGGKLNLPIYYRNKIFTEEERE 240
F GKVLCS GIG+GY KR DA +H Y G+T E+YR++NG K+NLPIYYRNK+FTE+ERE
Sbjct 181 FVGKVLCSRGIGAGYTKRPDAAKHKYKKGETIETYRLRNGAKINLPIYYRNKLFTEKERE 240
Query 241 KLFLDKIEKGIVYVLGSKIDLKTEESRYMGVLLSERERCE-RLYHDSPKDWDKRKYLNRL 299
LF+DKIEKGI+YV+G K+ EE Y LL E + E RLY + ++W+K+KYL+RL
Sbjct 241 LLFIDKIEKGIIYVMGQKVHRDDEE--YYLQLLDEGRKTECRLYGYNLQNWEKQKYLSRL 298
Query 300 KKQRQ 304
++Q++
Sbjct 299 RRQKK 303
> Alpavirinae_Human_feces_B_021_Microviridae_AG0372_putative.VP4
Length=332
Score = 383 bits (983), Expect = 1e-133, Method: Compositional matrix adjust.
Identities = 195/299 (65%), Positives = 237/299 (79%), Gaps = 1/299 (0%)
Query 1 MCLYPKLIPNKRYLPTKKNGGVPPVCPDERLRYVTAACGDCYECRKQKQRQWVVRMSEEN 60
MCLYPKLI NK+YLPTKKN PP D R Y+TAACG C ECRKQKQR+W+VRMSEE
Sbjct 1 MCLYPKLIRNKKYLPTKKNNYNPPKMADPRTAYITAACGKCLECRKQKQREWLVRMSEEL 60
Query 61 RQTPNAYFLTLTIddksykqlkqkyklkdnndIATKAIRLCLERVRKLTGKSVKHWFITE 120
R PNAYF+TLTI D++Y+ LK K +D N IATKAIRL LER+RK GKS++HWFITE
Sbjct 61 RTEPNAYFMTLTISDENYEILKNICKSEDENTIATKAIRLMLERIRKKIGKSIRHWFITE 120
Query 121 LGHEKTERLHLHGIVWGLGNGEKITNNWKYGITFTGYFVNEKTINYITKYMLKIDEKHPK 180
LGHEKTERLHLHGIVWG+G + I W YGIT+TG FVNEKTINYITKYM KIDE+HP
Sbjct 121 LGHEKTERLHLHGIVWGIGTDQLIREKWNYGITYTGNFVNEKTINYITKYMTKIDEEHPN 180
Query 181 FRGKVLCSAGIGSGYLKREDAKRHVYIPGKTNESYRMKNGGKLNLPIYYRNKIFTEEERE 240
F GKVLCS GIG+GY KR DA +H Y G+T E+YR++NG K+NLPIYYRNK+FTE+ERE
Sbjct 181 FVGKVLCSKGIGAGYTKRPDAAKHKYKKGETIETYRLRNGAKINLPIYYRNKLFTEKERE 240
Query 241 KLFLDKIEKGIVYVLGSKIDLKTEESRYMGVLLSERERCERLYHDSPKDWDKRKYLNRL 299
LF+DKI+KGI+YVLG+K+ + +E Y+ +L R++ LY + ++W+++KYLNRL
Sbjct 241 LLFIDKIDKGIIYVLGTKVH-RDDEKYYIQLLEEGRKKENMLYGNHTQEWEQQKYLNRL 298
> Alpavirinae_Human_feces_C_029_Microviridae_AG0108_putative.VP4
Length=291
Score = 349 bits (895), Expect = 7e-121, Method: Compositional matrix adjust.
Identities = 181/263 (69%), Positives = 221/263 (84%), Gaps = 3/263 (1%)
Query 43 ECRKQKQRQWVVRMSEENRQTPNAYFLTLTIddksykqlkqkyklkdnndIATKAIRLCL 102
ECR+QKQRQW+VRMSEE RQ PNAYF+TLTIDDKSY +L Y + DNN+IATKAIRLCL
Sbjct 2 ECRQQKQRQWLVRMSEELRQEPNAYFITLTIDDKSYSELSNTYNITDNNEIATKAIRLCL 61
Query 103 ERVRKLTGKSVKHWFITELGHEKTERLHLHGIVWGLGNGEKITNNWKYGITFTGYFVNEK 162
ER+RK TGKS+KHWFITELGHEKTERLHLHGIVWG+G + ITN W YGITFTG+FVNEK
Sbjct 62 ERIRKQTGKSIKHWFITELGHEKTERLHLHGIVWGIGTDKLITNKWNYGITFTGFFVNEK 121
Query 163 TINYITKYMLKIDEKHPKFRGKVLCSAGIGSGYLKREDAKRHVYIPGKTNESYRMKNGGK 222
TI YITKYM KIDE+H F GKVLCS GIG+GY+KR+DAK+H Y PG+T E+YR++NG K
Sbjct 122 TIQYITKYMTKIDEQHKDFIGKVLCSKGIGAGYIKRDDAKKHTYKPGETIETYRLRNGSK 181
Query 223 LNLPIYYRNKIFTEEEREKLFLDKIEKGIVYVLGSKIDLKTEESRYMGVLLSERERCE-R 281
+NLPIYYRN++FTEEE+E LFLDKIEKGI+YV+G K+ EE Y LL+E + E R
Sbjct 182 INLPIYYRNQLFTEEEKEALFLDKIEKGIIYVMGQKVHRDDEE--YYLQLLNEGRKTECR 239
Query 282 LYHDSPKDWDKRKYLNRLKKQRQ 304
LY + ++W+++KYL+RL++Q++
Sbjct 240 LYGYNLQNWEQQKYLSRLRRQKK 262
> Alpavirinae_Human_gut_31_037_Microviridae_AG0300_putative.VP4
Length=304
Score = 334 bits (856), Expect = 9e-115, Method: Compositional matrix adjust.
Identities = 177/304 (58%), Positives = 222/304 (73%), Gaps = 3/304 (1%)
Query 1 MCLYPKLIPNKRYLPTKKNGGVPPVCPDERLRYVTAACGDCYECRKQKQRQWVVRMSEEN 60
MCLYPKLI NKRY+PTKKN GV P CPDERLRYVTAACG CYECRKQK R W VR+SEE
Sbjct 1 MCLYPKLIKNKRYMPTKKNKGVVPPCPDERLRYVTAACGKCYECRKQKGRAWQVRLSEEV 60
Query 61 RQTPNAYFLTLTIddksykqlkqkyklkdnndIATKAIRLCLERVRKLTGKSVKHWFITE 120
R PNA F+TLTI D+S++++K Y + D K +RL LERVRK T KS+KHW TE
Sbjct 61 RSDPNAIFVTLTISDESWEKIKNTYIQLSDEDCIKKMVRLFLERVRKKTKKSLKHWLTTE 120
Query 121 LGHEKTERLHLHGIVWGLGNGEKITNN-WKYGITFTGYFVNEKTINYITKYMLKIDEKHP 179
G TER HLHG++WG N E +T + W+YG F G FVNE T+NYITKY+ K D+KH
Sbjct 121 RGGTNTERYHLHGLIWG-ENTEALTKSLWQYGFVFIGTFVNECTVNYITKYITKTDKKHK 179
Query 180 KFRGKVLCSAGIGSGYLKREDAKRHVYIPGKTNESYRMKNGGKLNLPIYYRNKIFTEEER 239
F LCSAGIG GYL R D++ + + GKT E+YR++NG KLNLPIYYRNK++T+EER
Sbjct 180 DFEPITLCSAGIGKGYLSRSDSELNRFREGKTTETYRLRNGTKLNLPIYYRNKLYTDEER 239
Query 240 EKLFLDKIEKGIVYVLGSKIDLKTEESRYMGVLLSERERCERLYHDSPKDWDKRKYLNRL 299
EKLFL+KI+KG V++ G + ++K ++ Y +L E+ + ++L+ DSP DWD KY RL
Sbjct 240 EKLFLEKIKKGKVWICGRECNIKDWKT-YSQILKEEQIKAKQLHGDSPIDWDIAKYERRL 298
Query 300 KKQR 303
+QR
Sbjct 299 ARQR 302
> Alpavirinae_Human_feces_A_032_Microviridae_AG0216_putative.VP4
Length=314
Score = 253 bits (645), Expect = 6e-83, Method: Compositional matrix adjust.
Identities = 147/310 (47%), Positives = 198/310 (64%), Gaps = 13/310 (4%)
Query 3 LYPKLIPNKRYLPTKKNGGVPPVCPDERLRYVTAACGDCYECRKQKQRQWVVRMSEENRQ 62
+Y K I NK+Y+ TKKN G P C DERLRYV CG C ECRK+K R W +R++EE +
Sbjct 1 MYTKYILNKKYMYTKKNQGNIPECRDERLRYVPTKCGKCIECRKEKARNWRIRLAEELKN 60
Query 63 TPNAYFLTLT--------IddksykqlkqkyklkdnndIATKAIRLCLERVRKLTGKSVK 114
PNA F+TLT + + +K+ K + N++ A+R LER+RK T KS++
Sbjct 61 NPNALFITLTFNEENYQRLSWELFKKSKDNLNYTEQNEMCKTAVRRWLERIRKKTKKSIR 120
Query 115 HWFITELGHEKTERLHLHGIVWGLGNGEKITNNWKYGITFTGYFVNEKTINYITKYMLKI 174
HW +TE G E R+HLHGIVW E+I W YG T+ G +VNE TI Y+TKYMLKI
Sbjct 121 HWMVTEKG-EDYGRIHLHGIVWC--PKERI-EQWGYGYTYIGDYVNETTIAYVTKYMLKI 176
Query 175 DEKHPKFRGKVLCSAGIGSGYLKREDAKRHVYIPGKTNESYRMKNGGKLNLPIYYRNKIF 234
EK P FRGKV+CSAGIG Y +AKR+ Y +T E+Y+++NG +L LP YY +KI+
Sbjct 177 CEKWPDFRGKVMCSAGIGCRYETSYNAKRNRYRGKETKETYKLENGRELPLPKYYHDKIY 236
Query 235 TEEEREKLFLDKIEKGIVYVLGSKIDLKTEESRYMGVLLSERERCERLYHDSPKDWDKRK 294
TEEEREKL++ K E+G Y+ G K+ E + + ++R E+LY D P+DW++ K
Sbjct 237 TEEEREKLWIIKQERGYRYIAGEKVSTDNLEE-WDNLTKYYQKRAEQLYGDKPEDWEREK 295
Query 295 YLNRLKKQRQ 304
RL+K RQ
Sbjct 296 TKARLEKMRQ 305
> Alpavirinae_Human_feces_B_023_Microviridae_AG0141_putative.VP4
Length=316
Score = 240 bits (613), Expect = 4e-78, Method: Compositional matrix adjust.
Identities = 139/311 (45%), Positives = 195/311 (63%), Gaps = 16/311 (5%)
Query 1 MCLYPKLIPNKRYLPTKKNGGVPPVCPDERLRYVTAACGDCYECRKQKQRQWVVRMSEEN 60
MCLY K I N +Y P KKN PPVC D RL YV CG C ECR+QKQR W+VR+SEE
Sbjct 1 MCLYTKYIENPKYKPNKKNNYNPPVCEDRRLFYVPVKCGKCIECRQQKQRAWIVRLSEEL 60
Query 61 RQTPNA-YFLTLTIddksykqlkqkyklkdnndIATKAIRLCLERV----RKLTGKSVKH 115
R A F+TLT +++SYK+L K RL L R+ R+ +++H
Sbjct 61 RSGKGAGLFVTLTFNEESYKELAAITKN------ENDMCRLALYRMNENYRQKYKHTIRH 114
Query 116 WFITELGHEKTERLHLHGIVWGLGNGEKITNNWKYGITFTGYFVNEKTINYITKYMLKID 175
W +TE+G + R+H+HGI+W + I WKYG + G FVNE+TI YITKYMLK
Sbjct 115 WCVTEIGDDG--RIHIHGIMWCPASD--IERYWKYGYIYIGRFVNEQTILYITKYMLKYT 170
Query 176 EKHPKFRGKVLCSAGIGSGYLKREDAKRHVYIPGKTNESYRMKNGGKLNLPIYYRNKIFT 235
F KVLCS GIG YL R D+KR+ Y T+ESY +++G K+NLP YY+ KI+T
Sbjct 171 PVDKNFEPKVLCSKGIGINYLDRLDSKRNTYRENNTDESYMLRSGRKINLPDYYKRKIYT 230
Query 236 EEEREKLFLDKIEKGIVYVLGSKIDLKTEESRYMGVLLSERERCERLYHDSPKDWDKRKY 295
EEEREKL+++K EKG Y++G K+ EE Y ++ R++ + LY++ P++WD+ K+
Sbjct 231 EEEREKLWIEKQEKGYRYIMGEKVSTDNEEKVY-KLMEYWRKKAKELYNEKPQEWDREKH 289
Query 296 LNRLKKQRQWI 306
+K++++++
Sbjct 290 KKAIKRRKEYL 300
> Alpavirinae_Human_feces_A_016_Microviridae_AG002_putative.VP4
Length=355
Score = 240 bits (612), Expect = 2e-77, Method: Compositional matrix adjust.
Identities = 150/346 (43%), Positives = 208/346 (60%), Gaps = 15/346 (4%)
Query 1 MCLYPKLIPNKRYLPTKKNGGVPPVCPDERLRYVTAACGDCYECRKQKQRQWVVRMSEEN 60
MC +P I NKR++PTKKNG PPVC DERLRY+ CG C+ECRK+K+ W VR E+
Sbjct 1 MCYFPIRIKNKRFVPTKKNGYEPPVCTDERLRYIEVECGYCFECRKKKRNAWRVRNFEQL 60
Query 61 RQTPNAYFLTLTIddksykqlkqkyklkdnndIATKAIRLCLERVRKLTGKSVKHWFITE 120
R+TP A F T T+ + Y +K+KY LK +N+I TK RL LER+RK TGKS+KHW +TE
Sbjct 61 RETPTAIFFTGTVSPERYDYIKEKYNLKTDNEIITKIHRLFLERIRKETGKSMKHWCVTE 120
Query 121 LGHEKTERLHLHGIVWGLGNGEK------ITNNWKYGITFTGYFVNEKTINYITKYMLKI 174
GH T R+HLHGI + + + NNW G + G + NEKTINY++KYM K
Sbjct 121 KGHTNTRRIHLHGIFYAPNGMTQFKLINILRNNWIDGYCYNGKYCNEKTINYVSKYMTKK 180
Query 175 DEKHPKFRGKVLCSAGIGSGYLKREDAKRHVYIPGKTNESYRMKNGGKLNLPIYYRNKIF 234
D +P++ GKVLCS G+G+GY+KR KRH + T E Y + G + LP YY+ K+F
Sbjct 181 DMDNPEYTGKVLCSPGLGAGYVKR-IGKRHEWNEENTKEDYYTRQGTYIALPKYYKYKLF 239
Query 235 TEEEREKLFLDKIEKGIVYVLGSKIDLKTEES-RYMGVLLSER-ERCERLYHDSPKDWDK 292
TE++RE+L++ + G +V K+ + EES Y VL + E R++ D K+
Sbjct 240 TEDQREQLWIYRENSGEKFVGNFKVKITDEESEEYYNVLKKQHNEDGIRIHKDDIKEIII 299
Query 293 RKYLNRLKKQRQWIESKATKVAekerkkeersekrLNNDIDLFANL 338
+K NR K +K +K + K++NNDI L ++
Sbjct 300 KKLQNRRDK------NKKSKAQRLFELYGKELRKKINNDIKLARDI 339
> Alpavirinae_Human_gut_30_040_Microviridae_AG0139_putative.VP4
Length=376
Score = 225 bits (573), Expect = 2e-71, Method: Compositional matrix adjust.
Identities = 148/324 (46%), Positives = 206/324 (64%), Gaps = 20/324 (6%)
Query 1 MCLYPKLIPNKRYLPTKKNGGVPPVCPDERLRYVTAACGDCYECRKQKQRQWVVRMSEEN 60
MCLYP I N +Y P KKN G PPVC D RL Y+ CG C ECRK+KQR+W VR+ EE
Sbjct 1 MCLYPTFIKNPKYKPNKKNKGNPPVCKDRRLLYIPVKCGCCIECRKEKQREWRVRLEEE- 59
Query 61 RQTPNAYFLTLTIddksykqlkqkyklkdnn---dIATKAIRLCLERVRKLTGKSVKHWF 117
+ YF TLTID ++ K+++ LK +IATKA+RL LER RK TGKS++HW
Sbjct 60 MHSNFGYFTTLTIDQENIKKIESITGLKWEENPNEIATKALRLFLERTRKDTGKSIRHWC 119
Query 118 ITELGHEKTERLHLHGIVWGLGNGEKITNNWKYGITFTGYFVNEKTINYITKYMLKIDEK 177
+TELG EK R+HLHGI +G + E I +W YG F G + N K+INYITKYMLK+D K
Sbjct 120 VTELG-EKNNRIHLHGIFFGQKSAELIRKHWNYGFIFIGGYCNSKSINYITKYMLKVDIK 178
Query 178 HPKFRGKVLCSAGIGSGYLKREDAKRHVYIPGKTN------ESYRMKNGGKLNLPIYYRN 231
HP+F+ VL S+GIG GY+ R D Y+ K N +Y +NG K+ +P YY+N
Sbjct 179 HPEFKQIVLASSGIGKGYIDRLD-----YLWQKQNYKNINVATYTFRNGTKMAMPKYYKN 233
Query 232 KIFTEEEREKLFLDKIEKGIVYVLGSKIDLKTEESRYMGVLLSERERCER-LYHDSPKDW 290
KIFTE+EREK++++ + +G++++ G K+ K ++ + L ++ R D+P W
Sbjct 234 KIFTEKEREKMWINNLNRGLLWIYGEKV--KADDWETIDNLREYWQKYGRETMGDNPIAW 291
Query 291 DKRKYLNRLKKQRQWIESKATKVA 314
+ K + +KQR+ I ++A K+A
Sbjct 292 NAMKERRKEEKQRRAI-AEAKKLA 314
Lambda K H a alpha
0.320 0.138 0.427 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 29743416