bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-20_CDS_annotation_glimmer3.pl_2_7
Length=606
Score E
Sequences producing significant alignments: (Bits) Value
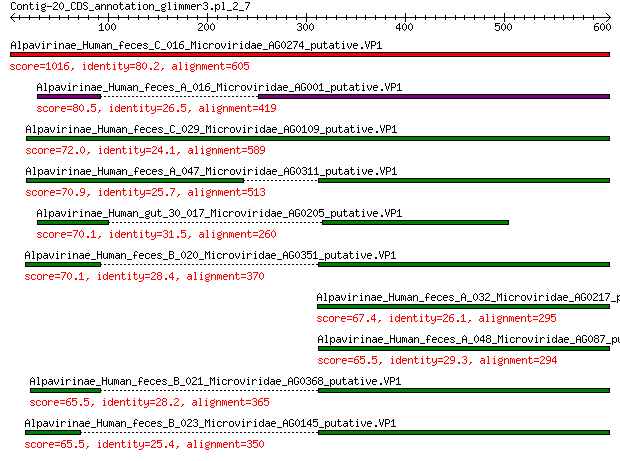
Alpavirinae_Human_feces_C_016_Microviridae_AG0274_putative.VP1 1016 0.0
Alpavirinae_Human_feces_A_016_Microviridae_AG001_putative.VP1 80.5 1e-17
Alpavirinae_Human_feces_C_029_Microviridae_AG0109_putative.VP1 72.0 5e-15
Alpavirinae_Human_feces_A_047_Microviridae_AG0311_putative.VP1 70.9 1e-14
Alpavirinae_Human_gut_30_017_Microviridae_AG0205_putative.VP1 70.1 2e-14
Alpavirinae_Human_feces_B_020_Microviridae_AG0351_putative.VP1 70.1 2e-14
Alpavirinae_Human_feces_A_032_Microviridae_AG0217_putative.VP1 67.4 1e-13
Alpavirinae_Human_feces_A_048_Microviridae_AG087_putative.VP1 65.5 5e-13
Alpavirinae_Human_feces_B_021_Microviridae_AG0368_putative.VP1 65.5 6e-13
Alpavirinae_Human_feces_B_023_Microviridae_AG0145_putative.VP1 65.5 6e-13
> Alpavirinae_Human_feces_C_016_Microviridae_AG0274_putative.VP1
Length=641
Score = 1016 bits (2626), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 485/607 (80%), Positives = 524/607 (86%), Gaps = 2/607 (0%)
Query 1 MSKYRIPSDYENQTPRAVHRRVASAYGTIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQP 60
MSKYRIPSDY+NQTPRA+HRR +SAYGTIHPGLA+PVHHRHLNVGERIRGRID LLQSQP
Sbjct 1 MSKYRIPSDYQNQTPRAMHRRASSAYGTIHPGLAIPVHHRHLNVGERIRGRIDELLQSQP 60
Query 61 MLGPLMNGYKLITIATFTPDSAIYGWMRNGRRYEPDEYTKFGKVYFPLAGSNIGNYIDPA 120
MLGPLMNG+KLITIATFTPDSAIYGWM NGRR+ PDEY KFGKVYF LAG N+ NY DPA
Sbjct 61 MLGPLMNGFKLITIATFTPDSAIYGWMSNGRRFTPDEYMKFGKVYFSLAGPNLENYKDPA 120
Query 121 FQIARRVRRLTFGLDLDANRAKIYESWVSDELNGTGDNGQPTHIGRGGLWDWLGIAPGAV 180
F+++R VRRLTFGLDL++++ KIYESWVSDELN TG +GQPTHIGRGGLWDWLGIA GAV
Sbjct 121 FKVSRPVRRLTFGLDLNSDQKKIYESWVSDELNATGSSGQPTHIGRGGLWDWLGIAAGAV 180
Query 181 CPNLGAKLSSVA-GIRGQLYPPSFKFNAAPFFAYFLSHYYYIANMQENYMYFTRGVGEMQ 239
CPNLG K +S + G+RG++YP SF+FNAAPF +YFLSHYYYIANMQE+YMYFTRGVGEMQ
Sbjct 181 CPNLGKKTTSSSPGVRGEIYPSSFQFNAAPFISYFLSHYYYIANMQEDYMYFTRGVGEMQ 240
Query 240 KVRPDGQQESLYRPFFSDVFTSFKPNEFLLYLDTLAYRTREGAQINAMDLKVEGLPI-NC 298
+VRPDGQQESLYRPFFSDVF S PN FL LD + TR G + LP N
Sbjct 241 RVRPDGQQESLYRPFFSDVFNSLDPNAFLYALDDVRNITRTGTGCELFKTFSDALPTSNS 300
Query 299 VQAMACAGIQGYGGLLSVPYSPDLFSNIIKQGSSPSIEITVENDQPNKPDAGFYVAVPEL 358
AMACAGIQGYGGLLSVPYSPDLF NIIKQGSSP+ EI V + D GF +AVPEL
Sbjct 301 FVAMACAGIQGYGGLLSVPYSPDLFGNIIKQGSSPTAEIEVIKALDSNTDTGFSIAVPEL 360
Query 359 RLKTKLQNWMDRLFVSGGRVGDVFRTLWGAKSSAPYVNKPDFLGVWQASVNPSNVRAMAN 418
RLKTK+QNWMDRLF+SGGRVGDVFRTLWG KSSAPYVNKPDFLGVWQAS+NPSNVRAMAN
Sbjct 361 RLKTKIQNWMDRLFISGGRVGDVFRTLWGTKSSAPYVNKPDFLGVWQASINPSNVRAMAN 420
Query 419 GSASGEDANLGQLAACIDRYCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYHQGLHPDLA 478
GSASGEDANLGQLAAC+DRYCDFS HSGIDYYAKEPGTFMLI MLVPEPAY QGLHPDLA
Sbjct 421 GSASGEDANLGQLAACVDRYCDFSDHSGIDYYAKEPGTFMLITMLVPEPAYSQGLHPDLA 480
Query 479 SVSFGDDFNPELNGIGFQSVPRHRFTMMPRGLDDITAHQEKNPWLGYAGTGVTIDPNTAS 538
S+SFGDDFNPELNGIGFQ VPRHRF+MMPRG + Q+ +PW G AGTGV IDPNT S
Sbjct 481 SISFGDDFNPELNGIGFQLVPRHRFSMMPRGFNLTGLDQQTSPWFGNAGTGVVIDPNTVS 540
Query 539 VGEEVAWSWLRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGFYGDAESYSTYINP 598
VGEEVAWSWLRTDY RLHGDFAQNGNYQYWVL R FT+YFPDAGTGFY D E TYINP
Sbjct 541 VGEEVAWSWLRTDYSRLHGDFAQNGNYQYWVLTRRFTTYFPDAGTGFYLDGEYTGTYINP 600
Query 599 LDWQYVF 605
LDWQYVF
Sbjct 601 LDWQYVF 607
> Alpavirinae_Human_feces_A_016_Microviridae_AG001_putative.VP1
Length=657
Score = 80.5 bits (197), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 93/365 (25%), Positives = 148/365 (41%), Gaps = 49/365 (13%)
Query 251 YRPFFSDVFTSFKPNEFLLYLDTLAYRTREGAQINAMDLKVEGLPI--NCVQAMACAGIQ 308
Y+ F + S E L LD + + N + L+ N + + A
Sbjct 300 YKKMFQESLGSILKEEKLEVLDKIRDEILKNPGNNILQLQTTTNTDVNNLFEDIRKASKN 359
Query 309 GYGGLLSVPYSPDLFSNIIK----QGSSPSIEITVENDQPNKPDAGFYVAVPELRLKTKL 364
GGL+ Y D+F+N +K G + ++T KP+ + + L L+ K+
Sbjct 360 KLGGLVIKTYDSDIFNNWVKTEWIDGENGISKVTAL-----KPNEDGTITMDALNLQQKV 414
Query 365 QNWMDRLFVSGGRVGDVFRTLWGAKSSAPYVNKPD---FLGVWQASVNPSNVRAMANGSA 421
N ++R+ VSGG D T++ A Y+++P+ F G + V A ++
Sbjct 415 YNMLNRIAVSGGTYRDWLETVYTA---GRYLDRPETPVFQGGMSQMIEFDEVVAT---TS 468
Query 422 SGEDANLGQLAA-CIDRYCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYHQGLHPDLASV 480
+ E LG+LA R + SG + + +EPG M + + P Y QG DL
Sbjct 469 NNEGQALGELAGRGYARQPNSSGR--LHFQVEEPGYVMGLVAITPMVDYSQGNDFDLNLF 526
Query 481 SFGDDFNPELNGIGFQSVPRHRFTMMPRGLDDITAHQEKNPWLGYAGTGVTIDPNTASVG 540
+ D P L+GIG+Q + E+ W G I T G
Sbjct 527 TMDDLHKPALDGIGYQDL-----------------MNEQRAWWTATQNGTKITDTTP--G 567
Query 541 EEVAWSWLRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGFYGDAESYSTYINPLD 600
+ VAW T++ R G+FA + + VL R +Y D G + STYINP +
Sbjct 568 KSVAWIDYMTNFNRTFGNFATGESEDFMVLNR---NYEDDEADGI----SNGSTYINPQE 620
Query 601 WQYVF 605
+F
Sbjct 621 HIDIF 625
Score = 26.6 bits (57), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 18/65 (28%), Positives = 28/65 (43%), Gaps = 2/65 (3%)
Query 28 TIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQPMLGPLMNGYKLITIATFTPDSAIY-GW 86
T+ G+ VP G+ ++ + + P LGPL +KL FT +Y W
Sbjct 39 TMGVGMLVPFMKILCQKGDIFDIKLTNKTMTHPTLGPLFGSFKLQHFI-FTAGFRLYNSW 97
Query 87 MRNGR 91
+ N R
Sbjct 98 LHNNR 102
> Alpavirinae_Human_feces_C_029_Microviridae_AG0109_putative.VP1
Length=640
Score = 72.0 bits (175), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 142/664 (21%), Positives = 215/664 (32%), Gaps = 156/664 (23%)
Query 17 AVHRRVASAYGTIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQPMLGPLMNGYKLITIAT 76
+ H T PG VP + G+ I I++ + + P +GPL +KL
Sbjct 26 STHDLSTVVRNTQAPGTLVPNLTLLVQKGDIIDIDIEANVLTHPTVGPLFGSFKLEHHVY 85
Query 77 FTPDSAIYGWMRNGRRYEPDEYTKFGKVYFPLAGSNIGNYIDPAFQIARRVRRLTFGLDL 136
P W+ N R TK G L
Sbjct 86 TVPFRLYNSWLHNNR-------TKIG---------------------------------L 105
Query 137 DANRAKIYESWVSDELN-GTGDNGQPTHIGRGGLWDWLGIAPGAVCPNLGAK-LSSVAGI 194
D ++ KI +LN G + PT +W I P + LG + S+ G+
Sbjct 106 DMSQVKI------PQLNVGLKNTDNPTSKN-----EWTQINPSCLLAYLGIRGYGSLIGV 154
Query 195 RGQLYPPSFKFNAAPFFAYFLSHYYYIANMQENYMYF----------------------- 231
+ NA P Y+ Y AN QE+ Y
Sbjct 155 TTKYVEK----NAVPLIGYYDIFKNYYANTQEDKFYTIGSGRKFTITINNVSYPITDINK 210
Query 232 TRGVGEMQKVRPD------------GQQESLYRPFF-SDVFTSFKPNEFLLYLDTLAYRT 278
T VG++ K P G + S YR + SD+ T + N+F + R
Sbjct 211 TLQVGDVIKFTPTPTKEEQGNLIFIGSENSRYRRWSGSDIGTWTEQNDFKISGLKAGKRY 270
Query 279 REGAQINAMDLKVEGLPINCVQAMACAGIQGYG-------GLLSVP-------------- 317
N + + ++ + + + + G G+ SVP
Sbjct 271 ELDRIFNELSVGLQQWELESLDTLRDNILTTKGNITFSVTGIDSVPVLQQFNERLGANSE 330
Query 318 ---------------YSPDLFSNIIKQGSSPSIEITVENDQPNKPDAGFYVAVPELRLKT 362
Y+ DL+ N I + E + D ++ L L
Sbjct 331 HLKTIGPQFGLALKTYNSDLYQNWINTDWIEGVNGINEISSVDVSDGNL--SMDALNLAQ 388
Query 363 KLQNWMDRLFVSGGRVGDVFRTLWGAKSSAPYVNKPDFLGVWQASVNPSNVRAMANGSAS 422
K+ N ++R+ VSGG D T++ P F G + V +++
Sbjct 389 KVYNMLNRIAVSGGTYRDWLETVFTGGEYMERCETPVFEGGTSQEI----VFQEVISNSA 444
Query 423 GEDANLGQLAA-CIDRYCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYHQGLHPDLASVS 481
E LG LA I GH I EPG M I + P Y QG D +
Sbjct 445 TEQEPLGTLAGRGITTNKQRGGH--IKIKVTEPGYIMCICSITPRIDYSQGNQWDTELKT 502
Query 482 FGDDFNPELNGIGFQSVPRHRFTMMPRGLDDITAHQEKNPWLGYAGTGVTIDPNTASVGE 541
D P L+GIG+Q + + E+ W GY D + G+
Sbjct 503 MDDLHKPALDGIGYQD----------------SINSERAWWAGYYNADP--DKKETAAGK 544
Query 542 EVAWSWLRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGFYGDAESYSTYINPLDW 601
VAW T+ R +G+FA N + V+ R + T STYI+P +
Sbjct 545 TVAWINYMTNVNRTYGNFAIKDNEAFMVMNRNYELEITGGSTPTKITIGDLSTYIDPAKF 604
Query 602 QYVF 605
Y+F
Sbjct 605 NYIF 608
> Alpavirinae_Human_feces_A_047_Microviridae_AG0311_putative.VP1
Length=637
Score = 70.9 bits (172), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 77/295 (26%), Positives = 114/295 (39%), Gaps = 27/295 (9%)
Query 312 GLLSVPYSPDLFSNIIKQGSSPSIEITVENDQPNKPDAGFYVAVPELRLKTKLQNWMDRL 371
GL Y+ DL+ N I + E + D ++ L L K+ N ++R+
Sbjct 337 GLALKTYNSDLYQNWINTDWIEGVNGINEISSVDVSDGSL--SMDALNLAQKVYNMLNRI 394
Query 372 FVSGGRVGDVFRTLWGAKSSAPYVNKPDFLGVWQASVNPSNVRAMANGSASGEDANLGQL 431
VSGG D T++ P F G + V + +++ E LG L
Sbjct 395 AVSGGTYRDWLETVFTGGEYMERCETPIFEGGTSQEIIFQEVIS----NSATEQEPLGTL 450
Query 432 AA-CIDRYCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYHQGLHPDLASVSFGDDFNPEL 490
A I GH I EPG M I + P Y QG D + D P L
Sbjct 451 AGRGITTNKQRGGHVKIK--VTEPGYIMCICSITPRIDYSQGNQWDTELKTMDDLHKPAL 508
Query 491 NGIGFQSVPRHRFTMMPRGLDDITAHQEKNPWLGYAGTGVTIDPNTASVGEEVAWSWLRT 550
+GIG+Q + + E+ W GY G + TA+ G+ VAW T
Sbjct 509 DGIGYQD----------------SINSERAWWAGYYSQG-PLKVETAA-GKTVAWINYMT 550
Query 551 DYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGFYGDAESYSTYINPLDWQYVF 605
+ R +G+FA N + V+ R + T STYI+P+ + Y+F
Sbjct 551 NVNRTYGNFAIKDNEAFMVMNRNYELQINGGPTPTSIRIGDLSTYIDPVKFNYIF 605
Score = 29.6 bits (65), Expect = 0.069, Method: Compositional matrix adjust.
Identities = 55/224 (25%), Positives = 75/224 (33%), Gaps = 63/224 (28%)
Query 17 AVHRRVASAYGTIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQPMLGPLMNGYKLITIAT 76
+ H T PG VP + G+ I I++ + + P +GPL +KL
Sbjct 26 STHDLSTVVRNTQAPGTLVPNLTLLVQKGDIIDIDIEANVLTHPTVGPLFGSFKLEHHIY 85
Query 77 FTPDSAIYGWMRNGRRYEPDEYTKFGKVYFPLAGSNIGNYIDPAFQIARRVRRLTFGLDL 136
P W+ N R TK G L
Sbjct 86 TIPFRLYNSWLHNNR-------TKIG---------------------------------L 105
Query 137 DANRAKIYESWVSDELNGTGDNGQPTHIGRGGLWDWLGIAPGAVCPNLGAKLSSVAGIR- 195
D ++ KI + ++ EL T DN PT +W + P + L GIR
Sbjct 106 DMSQVKIPQ--LNVELKNT-DN--PTTDN-----EWTQVNPSCLLAYL--------GIRG 147
Query 196 -GQLYPPSFKF---NAAPFFAYFLSHYYYIANMQENYMYFTRGV 235
G L S K+ NA P Y+ Y AN QE Y GV
Sbjct 148 YGSLIGTSAKYIGKNAVPLIGYYDIFKNYYANTQEEKFYTIGGV 191
> Alpavirinae_Human_gut_30_017_Microviridae_AG0205_putative.VP1
Length=647
Score = 70.1 bits (170), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 64/193 (33%), Positives = 84/193 (44%), Gaps = 24/193 (12%)
Query 316 VPYSPDLFSNIIKQGSSPSIEITVENDQPNKPDAGF--YVAVPELRLKTKLQNWMDRLFV 373
P SPD FS ++ G S S D F +P+L + T+LQ + D +
Sbjct 357 CPSSPDRFSRLMPPGDSNS-------------DVDFTGLKTIPQLAVATRLQEYKDLIGA 403
Query 374 SGGRVGDVFRTLWGAKSSAPYVNKPDFLGVWQASVNPSNV--RAMANGSASGEDANLGQL 431
SG R D T + +K +V++P L VN V +A +G A GE A LGQ+
Sbjct 404 SGSRYSDWLYTFFASK--IEHVDRPKLLFSSSVMVNSQVVMNQAGQSGFAGGEAAALGQM 461
Query 432 AACIDRYCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYH-QGLHPDLASVSFGDDFNPEL 490
I + G YY KEPG + ML P Y G+ PD D FNP
Sbjct 462 GGSI-AFNTVLGREQT-YYFKEPG--YIFDMLTIRPVYFWTGIRPDYLEYRGPDYFNPIY 517
Query 491 NGIGFQSVPRHRF 503
N IG+Q VP R
Sbjct 518 NDIGYQDVPLWRL 530
Score = 25.4 bits (54), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 18/73 (25%), Positives = 33/73 (45%), Gaps = 1/73 (1%)
Query 28 TIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQPMLGPLMNGYKLITIATFTPDSAIYGWM 87
+I PG+ PV + +N +R+ ++S P+ P N Y L + P + M
Sbjct 26 SITPGIIYPVRIQFVNARDRVTLHQGVDVRSNPLGVPSFNPYVLRLHRFWVPLQLYHPEM 85
Query 88 R-NGRRYEPDEYT 99
R N +++ + T
Sbjct 86 RVNSSKFDMNNLT 98
> Alpavirinae_Human_feces_B_020_Microviridae_AG0351_putative.VP1
Length=651
Score = 70.1 bits (170), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 85/302 (28%), Positives = 127/302 (42%), Gaps = 42/302 (14%)
Query 312 GLLSVPYSPDLFSNIIK----QGSSPSIEITVENDQPNKPDAGFYVAVPELRLKTKLQNW 367
GLL Y+ DL+ N I G++ EI+ + K + + L L K+ N
Sbjct 352 GLLLKTYNSDLYQNWINTDWIDGANGINEISSVDVSEGK------LTMDALNLAQKVYNM 405
Query 368 MDRLFVSGGRVGDVFRTLWGAKSSAPYVNKPDFLGVWQASVNPSNVRAMANGSASGEDAN 427
++R+ VSGG D T++ + P F G + V +++ ED
Sbjct 406 LNRIAVSGGTYRDWLETVYASGQYIERCETPTFEGGTSQEI----VFQEVVSNSATEDEP 461
Query 428 LGQLAACIDRYCDFSGHSG--IDYYAKEPGTFMLIAMLVPEPAYHQGLHPDLASVSFGDD 485
LG LA R + G I A EPG M I + P Y QG D + D
Sbjct 462 LGTLAG---RGVNAGKQKGGKIKVRATEPGYIMCITSITPRIDYSQGNDFDTDWKTLDDM 518
Query 486 FNPELNGIGFQ-SVPRHRFTMMPRGLDDI-TAHQEKNPWLGYAGTGVTIDPNTASVGEEV 543
P L+GIG+Q SV R DD+ T QE N + +TA G+ V
Sbjct 519 HKPALDGIGYQDSVNTGR-----AWWDDVYTGAQETN-----------LVKHTA--GKTV 560
Query 544 AWSWLRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGFYGDAESYSTYINPLDWQY 603
AW T+ + +G+FA + + VL R + + D+GT +TYI+P+ + Y
Sbjct 561 AWIDYMTNVNKTYGNFAAGMSEAFMVLNRNYEIKY-DSGTN--PRIADLTTYIDPIKYNY 617
Query 604 VF 605
+F
Sbjct 618 IF 619
Score = 33.5 bits (75), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 20/76 (26%), Positives = 28/76 (37%), Gaps = 0/76 (0%)
Query 16 RAVHRRVASAYGTIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQPMLGPLMNGYKLITIA 75
R+ H T PG VP + + I+S + + P GPL +KL
Sbjct 27 RSTHNLSTVIRNTQSPGTLVPTMTLVMQKDDTFDIEIESSVLTHPTTGPLYGSFKLENHL 86
Query 76 TFTPDSAIYGWMRNGR 91
F P W+ N R
Sbjct 87 FFIPFRLYNSWLHNNR 102
> Alpavirinae_Human_feces_A_032_Microviridae_AG0217_putative.VP1
Length=657
Score = 67.4 bits (163), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 77/307 (25%), Positives = 127/307 (41%), Gaps = 60/307 (20%)
Query 311 GGLLSVPYSPDLFSNIIKQ-------GSSPSIEITVENDQPNKPDAGFYVAVPELRLKTK 363
GGL Y DLF+N + + G + ++ V + N + L L K
Sbjct 367 GGLCLKTYQSDLFNNWVNKEWVDGDNGINAVTDVDVSEGKLN---------LDALNLAQK 417
Query 364 LQNWMDRLFVSGGRVGDVFRTLWGAKSSAPY---VNKPDFLGVWQASVNPSNVRAMANGS 420
+ N ++R+ +SGG D T++ + Y P + G +++ V + + +
Sbjct 418 VYNMLNRIAISGGSYKDWIETVY----TTDYYFRAETPVYEGGMSTTIDFEAVVSNSAST 473
Query 421 ASGEDANLGQLAACIDRYCDFSGHSG-IDYYAKEPGTFMLIAMLVPEPAYHQGLHPDLAS 479
ASG + LG LA R + + G I EP + IA + P Y QG D+
Sbjct 474 ASGIEEPLGSLAG---RGFNQGKNGGKIRIKCNEPCYIIGIASITPNVDYSQGNDWDMTQ 530
Query 480 VSFGDDFN-PELNGIGFQSVPRHRFTMMPRGLDDITAHQEKNPWLGYAGTGVTIDPNTAS 538
+ DD + P+L+GIG+Q + ++ + NP T +
Sbjct 531 LKTMDDLHKPQLDGIGYQDLLSNQMNGLA------------NP--------------TDA 564
Query 539 VGEEVAWSWLRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGFYGDAESYSTYINP 598
+G++ AW TD + + DFA Y VL R + + GT + STYI+P
Sbjct 565 IGKQPAWLNYMTDVNKTYADFAAGETESYMVLNRIY-DVNEETGTII-----NPSTYISP 618
Query 599 LDWQYVF 605
D+ Y+F
Sbjct 619 KDYTYIF 625
> Alpavirinae_Human_feces_A_048_Microviridae_AG087_putative.VP1
Length=650
Score = 65.5 bits (158), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 86/302 (28%), Positives = 122/302 (40%), Gaps = 43/302 (14%)
Query 312 GLLSVPYSPDLFSNIIK----QGSSPSIEITVENDQPNKPDAGFYVAVPELRLKTKLQNW 367
GL Y+ DL N I G + EI+ + K + + L L K+ N
Sbjct 352 GLCLKTYNSDLLQNWINTEWIDGVAGINEISAVDVTDGK------LTMDALNLSQKIYNM 405
Query 368 MDRLFVSGGRVGDVFRTLWGAKSSAPYVNKPDFLGVWQASVNPSNVRAMANGSASGEDAN 427
++R+ VSGG D T++ + P F G V + + + SASGE
Sbjct 406 LNRIAVSGGTYRDWLETVFTGGNYMERCETPMFEGGMSTEVV---FQEVISNSASGEQP- 461
Query 428 LGQLAACIDRYCDFSGHSG--IDYYAKEPGTFMLIAMLVPEPAYHQG--LHPDLASVSFG 483
LG LA R D G I EP M I + P Y QG +L +V
Sbjct 462 LGTLAG---RGYDTGKQKGGHIKIKVTEPCFIMGIGSITPRIDYSQGNEFFNELQTVD-- 516
Query 484 DDFNPELNGIGFQSVPRHRFTMMPRGLDDITAHQEKNPWLGYAGTGVTIDPNTASVGEEV 543
D P L+GIG+Q + R D T Q+ +S G+ V
Sbjct 517 DIHKPALDGIGYQDSLNWQ-----RAWWDDTRMQDNGRI-------------QSSAGKTV 558
Query 544 AWSWLRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGFYGDAESYSTYINPLDWQY 603
AW T+ R G+FA N N + VL R + P+AGT A+ +TYI+P+ + Y
Sbjct 559 AWINYMTNINRTFGNFAINDNEAFMVLNRNY-ELNPNAGTNETKIAD-LTTYIDPVKFNY 616
Query 604 VF 605
+F
Sbjct 617 IF 618
> Alpavirinae_Human_feces_B_021_Microviridae_AG0368_putative.VP1
Length=649
Score = 65.5 bits (158), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 85/302 (28%), Positives = 122/302 (40%), Gaps = 43/302 (14%)
Query 312 GLLSVPYSPDLFSNIIK----QGSSPSIEITVENDQPNKPDAGFYVAVPELRLKTKLQNW 367
GL Y+ DL N I G + EI+ + K + + L L K+ N
Sbjct 351 GLCLKTYNSDLLQNWINTEWIDGVTGINEISAVDVTDGK------LTMDALNLAQKVYNM 404
Query 368 MDRLFVSGGRVGDVFRTLWGAKSSAPYVNKPDFLGVWQASVNPSNVRAMANGSASGEDAN 427
++R+ VSGG D T++ + P F G + + + + SASGE
Sbjct 405 LNRIAVSGGTYRDWLETVFTGGNYMERCETPMFEGGMSTEIV---FQEVISNSASGEQP- 460
Query 428 LGQLAACIDRYCDFSGHSG--IDYYAKEPGTFMLIAMLVPEPAYHQG--LHPDLASVSFG 483
LG LA R D G I EP M I + P Y QG + +L +V
Sbjct 461 LGTLAG---RGYDTGKQKGGHIKIKVTEPCFIMGIGSITPRIDYSQGNEFYNELKTVD-- 515
Query 484 DDFNPELNGIGFQSVPRHRFTMMPRGLDDITAHQEKNPWLGYAGTGVTIDPNTASVGEEV 543
D P L+GIG+Q DD E N + +S G+ V
Sbjct 516 DIHKPALDGIGYQD----SLNWQRAWWDD--TRMENNGRI------------QSSAGKTV 557
Query 544 AWSWLRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGFYGDAESYSTYINPLDWQY 603
AW T+ R G+FA N N + VL R + P+AGT A+ +TYI+P+ + Y
Sbjct 558 AWINYMTNINRTFGNFAINDNEAFMVLNRNY-ELNPNAGTNETKIAD-LTTYIDPVKFNY 615
Query 604 VF 605
+F
Sbjct 616 IF 617
Score = 26.6 bits (57), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 18/71 (25%), Positives = 28/71 (39%), Gaps = 5/71 (7%)
Query 21 RVASAYGTIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQPMLGPLMNGYKLITIATFTPD 80
R + GT+ P L + G+ I I+S + + P +GPL +K P
Sbjct 37 RNTQSVGTLVPNLVLLAQK-----GDTIDIDIESHVLTHPTVGPLFGSFKHENHIFSVPI 91
Query 81 SAIYGWMRNGR 91
W+ N R
Sbjct 92 RLYNSWLHNNR 102
> Alpavirinae_Human_feces_B_023_Microviridae_AG0145_putative.VP1
Length=668
Score = 65.5 bits (158), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 74/301 (25%), Positives = 129/301 (43%), Gaps = 46/301 (15%)
Query 312 GLLSVPYSPDLFSNIIKQ----GSSPSIEITVENDQPNKPDAGFYVAVPELRLKTKLQNW 367
GLL Y D+F+N I G + IT + + NK + +L L K+ +
Sbjct 375 GLLLKTYQSDIFTNWINSEWIDGENGISAITAISTEGNK------FTIDQLNLSKKVYDM 428
Query 368 MDRLFVSGGRVGDVFRTLWGAKSSAPYVNKPDFLGVWQASVNPSNVRAMANGSASGEDAN 427
++R+ +SGG D T++ ++ + + P + G A + V + +++ E+
Sbjct 429 LNRIAISGGTYQDWVETVYTSEWNM-HTETPVYEGGMSAEIEFQEVVS----NSATEEEP 483
Query 428 LGQLAACIDRYCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYHQGLHPDLASVSFGDDFN 487
LG LA + + EP M IA + P Y QG D+ S+ DD +
Sbjct 484 LGTLAG--RGFASNKKGGQLHIKVTEPCYIMGIASITPRVDYCQGNDWDITSLDTMDDIH 541
Query 488 -PELNGIGFQSVPRHRFTMMPRGLDDITAHQEKNPWLGYAGTGVTIDPNTASVGEEVAWS 546
P+L+ IG+Q + + + + A +N +VG++ +W
Sbjct 542 KPQLDSIGYQDLMQEQ----------MNAQASRN----------------LAVGKQPSWI 575
Query 547 WLRTDYPRLHGDFA-QNGNYQ-YWVLARPFTSYFPDAGTGFYGDAESYSTYINPLDWQYV 604
T + + +G FA ++G + + VL R F DAGT + STYI+P + Y+
Sbjct 576 NYMTSFNKTYGTFANEDGEGEAFMVLNRYFDIKEIDAGTETGVKVYNTSTYIDPSQYNYI 635
Query 605 F 605
F
Sbjct 636 F 636
Score = 25.8 bits (55), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 15/56 (27%), Positives = 25/56 (45%), Gaps = 0/56 (0%)
Query 16 RAVHRRVASAYGTIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQPMLGPLMNGYKL 71
R+ H + ++ G VP G+ +ID+ + + P +GPL YKL
Sbjct 27 RSTHDLSYAWRSSMGVGTLVPCMKLLGLPGDTFDIKIDNKVLTHPTVGPLFGSYKL 82
Lambda K H a alpha
0.320 0.139 0.440 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 55802012