bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
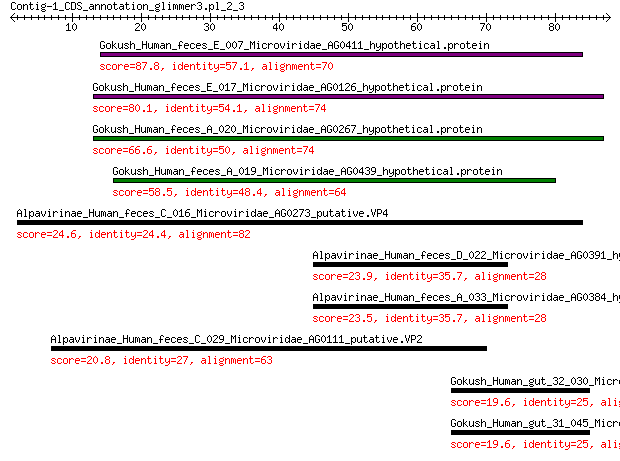
Query= Contig-1_CDS_annotation_glimmer3.pl_2_3
Length=87
Score E
Sequences producing significant alignments: (Bits) Value
Gokush_Human_feces_E_007_Microviridae_AG0411_hypothetical.protein 87.8 2e-25
Gokush_Human_feces_E_017_Microviridae_AG0126_hypothetical.protein 80.1 1e-22
Gokush_Human_feces_A_020_Microviridae_AG0267_hypothetical.protein 66.6 2e-17
Gokush_Human_feces_A_019_Microviridae_AG0439_hypothetical.protein 58.5 3e-14
Alpavirinae_Human_feces_C_016_Microviridae_AG0273_putative.VP4 24.6 0.11
Alpavirinae_Human_feces_D_022_Microviridae_AG0391_hypothetical.... 23.9 0.24
Alpavirinae_Human_feces_A_033_Microviridae_AG0384_hypothetical.... 23.5 0.34
Alpavirinae_Human_feces_C_029_Microviridae_AG0111_putative.VP2 20.8 3.0
Gokush_Human_gut_32_030_Microviridae_AG0187_putative.VP3 19.6 6.8
Gokush_Human_gut_31_045_Microviridae_AG0116_putative.VP3 19.6 6.8
> Gokush_Human_feces_E_007_Microviridae_AG0411_hypothetical.protein
Length=80
Score = 87.8 bits (216), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 40/70 (57%), Positives = 54/70 (77%), Gaps = 0/70 (0%)
Query 14 MNKTWNVRDQPRESLERQLEKKYKEIDGNYRMLRKVSNIEDAKMLVDEIWQMKSFANTIE 73
M+KTWNVRDQ E L + E+ YK+I+ Y+M++KVS+ EDAK ++D IW MK +AN IE
Sbjct 1 MHKTWNVRDQTEEGLRLEAERLYKQIEAGYKMIKKVSSSEDAKKIIDRIWIMKKWANDIE 60
Query 74 LELMRREYNN 83
LEL+RREY +
Sbjct 61 LELLRREYTH 70
> Gokush_Human_feces_E_017_Microviridae_AG0126_hypothetical.protein
Length=75
Score = 80.1 bits (196), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 40/74 (54%), Positives = 51/74 (69%), Gaps = 0/74 (0%)
Query 13 MMNKTWNVRDQPRESLERQLEKKYKEIDGNYRMLRKVSNIEDAKMLVDEIWQMKSFANTI 72
M+ K+WNVRDQ L Q EK Y I+ Y +++KVS IEDAK ++DEIW+MK +AN I
Sbjct 1 MIAKSWNVRDQTENELRFQEEKLYTNIEAKYMLIKKVSKIEDAKKIIDEIWRMKKWANDI 60
Query 73 ELELMRREYNNGTT 86
E+ELMRR N T
Sbjct 61 EIELMRRRANGEET 74
> Gokush_Human_feces_A_020_Microviridae_AG0267_hypothetical.protein
Length=75
Score = 66.6 bits (161), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 37/74 (50%), Positives = 48/74 (65%), Gaps = 0/74 (0%)
Query 13 MMNKTWNVRDQPRESLERQLEKKYKEIDGNYRMLRKVSNIEDAKMLVDEIWQMKSFANTI 72
M KTWNVRDQP E+L+++ EK Y I R + K + +E+AK LV+E + K+ AN I
Sbjct 1 MQVKTWNVRDQPTEALKKECEKCYTIIVKYNRWIAKAATMEEAKKLVEEKYAYKTKANDI 60
Query 73 ELELMRREYNNGTT 86
ELELMRR N T
Sbjct 61 ELELMRRRANGEET 74
> Gokush_Human_feces_A_019_Microviridae_AG0439_hypothetical.protein
Length=76
Score = 58.5 bits (140), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 31/64 (48%), Positives = 44/64 (69%), Gaps = 0/64 (0%)
Query 16 KTWNVRDQPRESLERQLEKKYKEIDGNYRMLRKVSNIEDAKMLVDEIWQMKSFANTIELE 75
KTWNVRDQ E L+++++K Y I R + + ++ E+AK L+DE + K+ AN IELE
Sbjct 5 KTWNVRDQTTEVLKKEVDKCYTMIVKYNRWIARAASAEEAKSLIDEKFNYKAKANNIELE 64
Query 76 LMRR 79
LMRR
Sbjct 65 LMRR 68
> Alpavirinae_Human_feces_C_016_Microviridae_AG0273_putative.VP4
Length=304
Score = 24.6 bits (52), Expect = 0.11, Method: Composition-based stats.
Identities = 20/90 (22%), Positives = 38/90 (42%), Gaps = 13/90 (14%)
Query 2 ITSTKMKRSKAMMNKTWNVRDQPRESLERQLEKKYKEIDGNYRMLRKVSNIEDAKMLVDE 61
I S + ++ ++K N R QP L +++ Y + S+++ M++D
Sbjct 184 IGSNYLDTEESSLHKLGNQRYQPFMVL-----NGFQQAMPRYYYNKLFSDVDKQNMVIDR 238
Query 62 I--------WQMKSFANTIELELMRREYNN 83
WQ + F + +E + MRR N
Sbjct 239 FINPPVEFSWQGQKFGSKLERDEMRRSTLN 268
> Alpavirinae_Human_feces_D_022_Microviridae_AG0391_hypothetical.protein.BACEGG.02723
Length=367
Score = 23.9 bits (50), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 10/28 (36%), Positives = 16/28 (57%), Gaps = 0/28 (0%)
Query 45 MLRKVSNIEDAKMLVDEIWQMKSFANTI 72
+ R ++NI D K + D IW++ S I
Sbjct 319 LARYLANIHDPKNMWDGIWRLVSLPAGI 346
> Alpavirinae_Human_feces_A_033_Microviridae_AG0384_hypothetical.protein.BACEGG.02723
Length=367
Score = 23.5 bits (49), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 10/28 (36%), Positives = 16/28 (57%), Gaps = 0/28 (0%)
Query 45 MLRKVSNIEDAKMLVDEIWQMKSFANTI 72
+ R ++NI D K + D IW++ S I
Sbjct 319 LARYLANIHDPKNMWDGIWRIVSLPAGI 346
> Alpavirinae_Human_feces_C_029_Microviridae_AG0111_putative.VP2
Length=329
Score = 20.8 bits (42), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 17/63 (27%), Positives = 31/63 (49%), Gaps = 12/63 (19%)
Query 7 MKRSKAMMNKTWNVRDQPRESLERQLEKKYKEIDGNYRMLRKVSNIEDAKMLVDEIWQMK 66
+K KA++N+ RDQ +E +++ Y + +K +IE+ K++ D I M
Sbjct 261 VKEGKALINQ----RDQ--------IEAYVQDVINRYELGKKGLDIEEQKLVKDVILGML 308
Query 67 SFA 69
A
Sbjct 309 EIA 311
> Gokush_Human_gut_32_030_Microviridae_AG0187_putative.VP3
Length=157
Score = 19.6 bits (39), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 5/20 (25%), Positives = 14/20 (70%), Gaps = 0/20 (0%)
Query 65 MKSFANTIELELMRREYNNG 84
++S+A + ++ ++ + Y NG
Sbjct 49 IQSYAESCDIHVLMKRYANG 68
> Gokush_Human_gut_31_045_Microviridae_AG0116_putative.VP3
Length=157
Score = 19.6 bits (39), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 5/20 (25%), Positives = 14/20 (70%), Gaps = 0/20 (0%)
Query 65 MKSFANTIELELMRREYNNG 84
++S+A + ++ ++ + Y NG
Sbjct 49 IQSYAESCDIHVLMKRYANG 68
Lambda K H a alpha
0.313 0.125 0.345 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3840903