bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-16_CDS_annotation_glimmer3.pl_2_5
Length=168
Score E
Sequences producing significant alignments: (Bits) Value
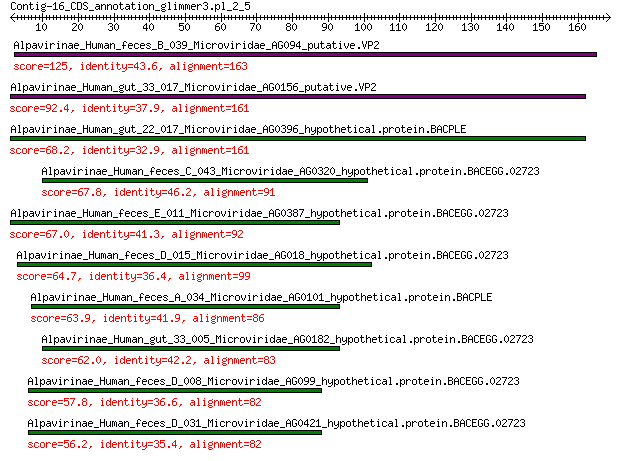
Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2 125 3e-36
Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2 92.4 2e-24
Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.p... 68.2 1e-15
Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.... 67.8 2e-15
Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.... 67.0 3e-15
Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.p... 64.7 2e-14
Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.... 63.9 3e-14
Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.p... 62.0 1e-13
Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.p... 57.8 4e-12
Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.... 56.2 1e-11
> Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2
Length=380
Score = 125 bits (315), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 71/163 (44%), Positives = 108/163 (66%), Gaps = 0/163 (0%)
Query 2 EFAGRLTSAQEAQILLDSEAQQVLNKYLDEQQQADLFIKGQTLSNLYAQGSLTEAQYKTE 61
+FA R+++AQEAQILL+S+AQ+++NKY+D+ QQADLFIK QTL+NL +QG+LTE Q +TE
Sbjct 194 QFAERISAAQEAQILLNSDAQRIMNKYMDQNQQADLFIKAQTLANLQSQGALTEKQIQTE 253
Query 62 MAKAVKTAAETNGIRINNKIASQTADSLIYANIQSNRARGLSSLWDSKNTNVLKNIEYSK 121
+ +A+ +AE +G +I+N++AS+TADSLI A SN + S +D KN + K+ EY
Sbjct 254 IQRAILASAEASGKKIDNRVASETADSLIKAANASNELQYRDSTYDYKNVKLRKHTEYKT 313
Query 122 DKALRDYYKWSSKHKQKDVDSYELRNAVDYGTRIFQGIGNSIG 164
A + ++ + +K ++ + I G GN IG
Sbjct 314 SMANQKAAEYGADLARKQGRTHYWESVARGIGSIAAGAGNFIG 356
> Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2
Length=353
Score = 92.4 bits (228), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 61/161 (38%), Positives = 91/161 (57%), Gaps = 7/161 (4%)
Query 1 MEFAGRLTSAQEAQILLDSEAQQVLNKYLDEQQQADLFIKGQTLSNLYAQGSLTEAQYKT 60
M FAGRL AQ LL+++A+ VLN+YLD+QQQADL +K N +QG L Q K
Sbjct 196 MAFAGRLLQAQGTSQLLEADAKTVLNRYLDQQQQADLNVKASVYYNQMSQGHLNYNQAKK 255
Query 61 EMAKAVKTAAETNGIRINNKIASQTADSLIYANIQSNRARGLSSLWDSKNTNVLKNIEYS 120
+A + T A G +++NK+A TADSLI A +NR+ L +K N E +
Sbjct 256 VIADEILTYARIKGQKLSNKVAEATADSLIRATNAANRSNAEFDLEAAK-----YNRERA 310
Query 121 KDKALRDYYKWSSKHKQKDVDSYELRNAVDYGTRIFQGIGN 161
+ +++ D+Y+ S+++ K Y+ + YGT I +GN
Sbjct 311 RSRSIEDWYR--SRNEGKKYKYYDSDKLIHYGTSIGNTVGN 349
> Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.protein.BACPLE
Length=386
Score = 68.2 bits (165), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 53/161 (33%), Positives = 92/161 (57%), Gaps = 5/161 (3%)
Query 1 MEFAGRLTSAQEAQILLDSEAQQVLNKYLDEQQQADLFIKGQTLSNLYAQGSLTEAQYKT 60
M G + AQ + +LLD+EA+ +LNKYLD+ QQ DL +K A G ++ A+ K
Sbjct 196 MAMTGLVLRAQRSGMLLDNEAKGILNKYLDQHQQLDLSVKAADYYQRMAAGYVSYAEAKK 255
Query 61 EMAKAVKTAAETNGIRINNKIASQTADSLIYANIQSNRARGLSSLWDSKNTNVLKNIEYS 120
+A+ AA T G I+N++AS+ A+S I ANI +N + S+ + ++ + + +
Sbjct 256 ALAEEALAAARTRGQNISNEVASRIAESQIAANIAANES---SAAYHNEELRLGLPQDNA 312
Query 121 KDKALRDYYKWSSKHKQKDVDSYELRNAVDYGTRIFQGIGN 161
+ K + ++Y+ S++++K ++ V+YGT I IGN
Sbjct 313 RSKNIEEWYR--SRNEKKRYKYFDADKWVEYGTSIGNTIGN 351
> Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.protein.BACEGG.02723
Length=396
Score = 67.8 bits (164), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 42/91 (46%), Positives = 57/91 (63%), Gaps = 0/91 (0%)
Query 10 AQEAQILLDSEAQQVLNKYLDEQQQADLFIKGQTLSNLYAQGSLTEAQYKTEMAKAVKTA 69
A+ A L ++AQ+ LNKYLD QQ L K S++ A LT+A+Y+TE+A +KT
Sbjct 202 AETAVASLTADAQRTLNKYLDMGQQLSLITKMAEYSSITAGTELTKAKYRTEIANEIKTL 261
Query 70 AETNGIRINNKIASQTADSLIYANIQSNRAR 100
AE NG +I+N+IA TA SLI A + N R
Sbjct 262 AEANGQKISNEIARSTAQSLIDAMNKENEYR 292
> Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.protein.BACEGG.02723
Length=410
Score = 67.0 bits (162), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 38/92 (41%), Positives = 55/92 (60%), Gaps = 0/92 (0%)
Query 1 MEFAGRLTSAQEAQILLDSEAQQVLNKYLDEQQQADLFIKGQTLSNLYAQGSLTEAQYKT 60
M++ ++ AQ ILL ++A+ ++NKYLD Q L + +A G L+ Q KT
Sbjct 239 MKWVNKIQRAQRTDILLSNQAKGIINKYLDTSQSLQLKLMANQSFQAFASGRLSLQQCKT 298
Query 61 EMAKAVKTAAETNGIRINNKIASQTADSLIYA 92
E+ K + AET G +I+NKIAS+TAD LI A
Sbjct 299 EVTKQLMNMAETEGKKISNKIASETADQLIGA 330
> Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.protein.BACEGG.02723
Length=427
Score = 64.7 bits (156), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 36/99 (36%), Positives = 58/99 (59%), Gaps = 0/99 (0%)
Query 3 FAGRLTSAQEAQILLDSEAQQVLNKYLDEQQQADLFIKGQTLSNLYAQGSLTEAQYKTEM 62
++ L A A LLD+E++ +LNKYLD+QQQADL +K L +G L + + +
Sbjct 225 WSNNLLRANVANSLLDAESKTILNKYLDQQQQADLNVKAAHYEELINRGQLHVVEARELL 284
Query 63 AKAVKTAAETNGIRINNKIASQTADSLIYANIQSNRARG 101
++ V A G+ I+N +A+++A L+YAN +N G
Sbjct 285 SREVLNYARARGLNISNWVAAKSAKGLVYANNAANYYEG 323
> Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.protein.BACPLE
Length=365
Score = 63.9 bits (154), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 36/86 (42%), Positives = 53/86 (62%), Gaps = 0/86 (0%)
Query 7 LTSAQEAQILLDSEAQQVLNKYLDEQQQADLFIKGQTLSNLYAQGSLTEAQYKTEMAKAV 66
+ +AQ+A +LL +EAQQV+N YL +++Q L G N+ GS+ E Q K +A +
Sbjct 175 VATAQKANLLLSAEAQQVMNMYLPQEKQIQLSTLGAQYWNMIRDGSIKEEQAKNLLATRL 234
Query 67 KTAAETNGIRINNKIASQTADSLIYA 92
+ A T G I+NK+A TADS+I A
Sbjct 235 EIEARTAGQHISNKVARSTADSIIDA 260
> Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.protein.BACEGG.02723
Length=354
Score = 62.0 bits (149), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 53/83 (64%), Gaps = 0/83 (0%)
Query 10 AQEAQILLDSEAQQVLNKYLDEQQQADLFIKGQTLSNLYAQGSLTEAQYKTEMAKAVKTA 69
AQ+A +LL ++AQ+VLN YL E+++ L + G N+ +G ++E Q K +A ++
Sbjct 178 AQKANLLLRNDAQEVLNMYLPEEKRIQLQMNGAQYWNMIREGVISEEQAKNLIASRLEIE 237
Query 70 AETNGIRINNKIASQTADSLIYA 92
A T G I+NKIA TADS+I A
Sbjct 238 ARTQGQHISNKIAKSTADSIIDA 260
> Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.protein.BACEGG.02723
Length=418
Score = 57.8 bits (138), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 30/82 (37%), Positives = 51/82 (62%), Gaps = 0/82 (0%)
Query 6 RLTSAQEAQILLDSEAQQVLNKYLDEQQQADLFIKGQTLSNLYAQGSLTEAQYKTEMAKA 65
++ AQ A + L +E Q+++NK++ QQQA+ F+K Y G L+EAQ KT++ +
Sbjct 222 KVALAQGALLGLQAEGQRIVNKFMPAQQQAEFFLKTANAFAQYKAGKLSEAQVKTQIKQQ 281
Query 66 VKTAAETNGIRINNKIASQTAD 87
A+T G ++NN++A + AD
Sbjct 282 ALLEAQTVGQKLNNRLAERLAD 303
> Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.protein.BACEGG.02723
Length=418
Score = 56.2 bits (134), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 29/82 (35%), Positives = 50/82 (61%), Gaps = 0/82 (0%)
Query 6 RLTSAQEAQILLDSEAQQVLNKYLDEQQQADLFIKGQTLSNLYAQGSLTEAQYKTEMAKA 65
++ AQ A + L +E Q+++NK++ QQQA+ F+K Y G L+EAQ KT++ +
Sbjct 222 KVALAQGALLGLQAEGQRIVNKFMPAQQQAEFFLKTANAFAQYKAGKLSEAQVKTQIKQQ 281
Query 66 VKTAAETNGIRINNKIASQTAD 87
A+ G ++NN++A + AD
Sbjct 282 ALLEAQAAGQKLNNRLAERLAD 303
Lambda K H a alpha
0.310 0.125 0.337 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 11677680