bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-10_CDS_annotation_glimmer3.pl_2_1
Length=279
Score E
Sequences producing significant alignments: (Bits) Value
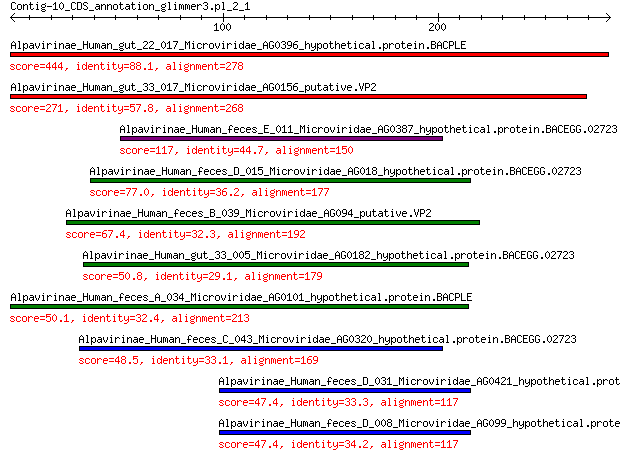
Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.p... 444 1e-157
Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2 271 1e-90
Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.... 117 5e-32
Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.p... 77.0 7e-18
Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2 67.4 1e-14
Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.p... 50.8 3e-09
Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.... 50.1 6e-09
Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.... 48.5 2e-08
Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.... 47.4 5e-08
Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.p... 47.4 6e-08
> Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.protein.BACPLE
Length=386
Score = 444 bits (1141), Expect = 1e-157, Method: Compositional matrix adjust.
Identities = 245/278 (88%), Positives = 264/278 (95%), Gaps = 0/278 (0%)
Query 1 MMNGGSAGVaqsagngataassgnaVMQPFQADYSGIGSSIGNIFQYDLMQSEKSQLQGA 60
MMNGGSAGVAQSAG G+ A+SSGNAVMQPFQADYSGIGSSIGNIFQY+LMQSEKSQLQGA
Sbjct 87 MMNGGSAGVAQSAGTGSAASSSGNAVMQPFQADYSGIGSSIGNIFQYELMQSEKSQLQGA 146
Query 61 RQLSDAKAMETLSNIDWGNLTAETRNYLRSTGLARAQLGYVKEQQEVDNMAMTGLIMRAQ 120
RQL+DAKAMETLSNIDWG LT ETR +L+STGLARAQLGY KEQQE DNMAMTGL++RAQ
Sbjct 147 RQLADAKAMETLSNIDWGKLTDETRGFLKSTGLARAQLGYAKEQQEADNMAMTGLVLRAQ 206
Query 121 RSGMLLDNEAKGILNKYLDQEQQLDLNVKAADYYQRMSAGYLSYaetkkaladealaaar 180
RSGMLLDNEAKGILNKYLDQ QQLDL+VKAADYYQRM+AGY+SYAE KKALA+EALAAAR
Sbjct 207 RSGMLLDNEAKGILNKYLDQHQQLDLSVKAADYYQRMAAGYVSYAEAKKALAEEALAAAR 266
Query 181 trGQKISNEVASRIAESQIAANIAANQSSEAYHNEELKLGLSQDNARSKNIEEWYRSRNE 240
TRGQ ISNEVASRIAESQIAANIAAN+SS AYHNEEL+LGL QDNARSKNIEEWYRSRNE
Sbjct 267 TRGQNISNEVASRIAESQIAANIAANESSAAYHNEELRLGLPQDNARSKNIEEWYRSRNE 326
Query 241 KKRYKYYDADKWVEYGTNIGNIIGNFLPRRIVSKTFPR 278
KKRYKY+DADKWVEYGT+IGN IGNFLPRR++SK+F R
Sbjct 327 KKRYKYFDADKWVEYGTSIGNTIGNFLPRRVISKSFSR 364
> Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2
Length=353
Score = 271 bits (693), Expect = 1e-90, Method: Compositional matrix adjust.
Identities = 155/268 (58%), Positives = 209/268 (78%), Gaps = 2/268 (1%)
Query 1 MMNGGSAGVaqsagngataassgnaVMQPFQADYSGIGSSIGNIFQYDLMQSEKSQLQGA 60
MMNGGS+GV+QSAG GA+A+SSG AV QPFQAD+SGI +IG++FQ + Q++ SQ+QG
Sbjct 87 MMNGGSSGVSQSAGTGASASSSGTAVFQPFQADFSGIQQAIGSVFQSQVRQAQVSQMQGQ 146
Query 61 RQLSDAKAMETLSNIDWGNLTAETRNYLRSTGLARAQLGYVKEQQEVDNMAMTGLIMRAQ 120
R L+DA+AM+ LS +DW +T ETR YL++TGLARA+LGY KE QE+DNMA G +++AQ
Sbjct 147 RNLADAQAMQALSQVDWSKMTKETREYLKATGLARARLGYSKEMQELDNMAFAGRLLQAQ 206
Query 121 RSGMLLDNEAKGILNKYLDQEQQLDLNVKAADYYQRMSAGYLSYaetkkaladealaaar 180
+ LL+ +AK +LN+YLDQ+QQ DLNVKA+ YY +MS G+L+Y + KK +ADE L AR
Sbjct 207 GTSQLLEADAKTVLNRYLDQQQQADLNVKASVYYNQMSQGHLNYNQAKKVIADEILTYAR 266
Query 181 trGQKISNEVASRIAESQIAANIAANQSSEAYHNEELKLGLSQDNARSKNIEEWYRSRNE 240
+GQK+SN+VA A+S I A AAN+S+ + E K +++ ARS++IE+WYRSRNE
Sbjct 267 IKGQKLSNKVAEATADSLIRATNAANRSNAEFDLEAAK--YNRERARSRSIEDWYRSRNE 324
Query 241 KKRYKYYDADKWVEYGTNIGNIIGNFLP 268
K+YKYYD+DK + YGT+IGN +GNFLP
Sbjct 325 GKKYKYYDSDKLIHYGTSIGNTVGNFLP 352
> Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.protein.BACEGG.02723
Length=410
Score = 117 bits (294), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 67/150 (45%), Positives = 96/150 (64%), Gaps = 0/150 (0%)
Query 52 SEKSQLQGARQLSDAKAMETLSNIDWGNLTAETRNYLRSTGLARAQLGYVKEQQEVDNMA 111
+E + LQG + L+DA+A TLS IDW T E R +L++TG+ARAQL + +QQ ++NM
Sbjct 181 AEANNLQGQKGLADAQAAATLSGIDWYKFTPEYRAWLQTTGMARAQLSFNTDQQNLENMK 240
Query 112 MTGLIMRAQRSGMLLDNEAKGILNKYLDQEQQLDLNVKAADYYQRMSAGYLSYaetkkal 171
I RAQR+ +LL N+AKGI+NKYLD Q L L + A +Q ++G LS + K +
Sbjct 241 WVNKIQRAQRTDILLSNQAKGIINKYLDTSQSLQLKLMANQSFQAFASGRLSLQQCKTEV 300
Query 172 adealaaartrGQKISNEVASRIAESQIAA 201
+ + A T G+KISN++AS A+ I A
Sbjct 301 TKQLMNMAETEGKKISNKIASETADQLIGA 330
> Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.protein.BACEGG.02723
Length=427
Score = 77.0 bits (188), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 64/177 (36%), Positives = 97/177 (55%), Gaps = 14/177 (8%)
Query 38 GSSIGNIFQYDLMQSEKSQLQGARQLSDAKAMETLSNIDWGNLTAETRNYLRSTGLARAQ 97
G I N D +QS+ +++G + +W N++ E Y +GL A+
Sbjct 165 GLDIRNFSLKDYLQSQIDKMKG--------------DTNWRNVSPEAIRYNIMSGLEAAK 210
Query 98 LGYVKEQQEVDNMAMTGLIMRAQRSGMLLDNEAKGILNKYLDQEQQLDLNVKAADYYQRM 157
+G +++ N + ++RA + LLD E+K ILNKYLDQ+QQ DLNVKAA Y + +
Sbjct 211 IGMENLREQWANQVWSNNLLRANVANSLLDAESKTILNKYLDQQQQADLNVKAAHYEELI 270
Query 158 SAGYLSYaetkkaladealaaartrGQKISNEVASRIAESQIAANIAANQSSEAYHN 214
+ G L E ++ L+ E L AR RG ISN VA++ A+ + AN AAN +Y+N
Sbjct 271 NRGQLHVVEARELLSREVLNYARARGLNISNWVAAKSAKGLVYANNAANYYEGSYNN 327
> Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2
Length=380
Score = 67.4 bits (163), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 62/207 (30%), Positives = 102/207 (49%), Gaps = 20/207 (10%)
Query 27 MQPFQADYSGIGSSIGNIFQYDLMQSEKSQLQGARQLSDAKAMETLSNI---------DW 77
MQ ++ ++S + S+ + L Q++ S+ S A+ +T++ + +W
Sbjct 105 MQAYKPNFSSVFQSLAS-----LAQAKASEASAGESGSRARQTDTVTPLLSDYYRGLTNW 159
Query 78 GNLTAETRNYL-RSTGLARAQLGYVKEQQEVDNMAMTGLIMRAQRSGMLLDNEAKGILNK 136
NL + Y + TG A L E Q + N I AQ + +LL+++A+ I+NK
Sbjct 160 KNLAIGSSGYWNKETGRVSAALDQSTETQNLKNAQFAERISAAQEAQILLNSDAQRIMNK 219
Query 137 YLDQEQQLDLNVKAADYYQRMSAGYLSYaetkkaladealaaartrGQKISNEVASRIAE 196
Y+DQ QQ DL +KA S G L+ + + + LA+A G+KI N VAS A+
Sbjct 220 YMDQNQQADLFIKAQTLANLQSQGALTEKQIQTEIQRAILASAEASGKKIDNRVASETAD 279
Query 197 SQIAANIAANQ-----SSEAYHNEELK 218
S I A A+N+ S+ Y N +L+
Sbjct 280 SLIKAANASNELQYRDSTYDYKNVKLR 306
> Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.protein.BACEGG.02723
Length=354
Score = 50.8 bits (120), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 52/184 (28%), Positives = 91/184 (49%), Gaps = 19/184 (10%)
Query 35 SGIGSSIGNIFQY---DLMQSEKSQLQGARQLSDAKAMETLSNI--DWGNLTAETRNYLR 89
SG S G +QY +++ S + +SDA+ T S++ +G T E++
Sbjct 103 SGASGSGGTPYQYTPTNMIGDVASYASAMKSMSDARKTNTESDLLDKYGVPTYESQ---- 158
Query 90 STGLARAQLGYVKEQQEVDNMAMTGLIMRAQRSGMLLDNEAKGILNKYLDQEQQLDLNVK 149
G A + + Q +V AQ++ +LL N+A+ +LN YL +E+++ L +
Sbjct 159 -IGKTMADTYFTQRQADV---------AIAQKANLLLRNDAQEVLNMYLPEEKRIQLQMN 208
Query 150 AADYYQRMSAGYLSYaetkkaladealaaartrGQKISNEVASRIAESQIAANIAANQSS 209
A Y+ + G +S + K +A ART+GQ ISN++A A+S I A A ++
Sbjct 209 GAQYWNMIREGVISEEQAKNLIASRLEIEARTQGQHISNKIAKSTADSIIDATRTAKENE 268
Query 210 EAYH 213
A++
Sbjct 269 AAFN 272
> Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.protein.BACPLE
Length=365
Score = 50.1 bits (118), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 69/216 (32%), Positives = 108/216 (50%), Gaps = 31/216 (14%)
Query 1 MMNGGSAGVaqsagngataassgnaVMQPFQADYSGIGSSIGNIFQYDLMQSEKSQLQGA 60
MMNGGSAG AQS A++ SSG+ M P+Q + + IG++ Y GA
Sbjct 85 MMNGGSAGTAQSTSATASSGSSGSGGM-PYQYTPTNV---IGDVASY----------AGA 130
Query 61 -RQLSDAKAMETLSNI--DWGNLTAETRNYLRSTGLARAQLGYVKEQQEVDNMAMTGLIM 117
+ LSDA+ T +++ +G+ +R +A + +Q++ D +
Sbjct 131 MKSLSDARKSGTEADLLGRYGDSDYSSR-------IANTEADTYFKQRQSD-------VA 176
Query 118 RAQRSGMLLDNEAKGILNKYLDQEQQLDLNVKAADYYQRMSAGYLSYaetkkaladeala 177
AQ++ +LL EA+ ++N YL QE+Q+ L+ A Y+ + G + + K LA
Sbjct 177 TAQKANLLLSAEAQQVMNMYLPQEKQIQLSTLGAQYWNMIRDGSIKEEQAKNLLATRLEI 236
Query 178 aartrGQKISNEVASRIAESQIAANIAANQSSEAYH 213
ART GQ ISN+VA A+S I A A + AY+
Sbjct 237 EARTAGQHISNKVARSTADSIIDATNTAKMNEAAYN 272
> Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.protein.BACEGG.02723
Length=396
Score = 48.5 bits (114), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 56/169 (33%), Positives = 80/169 (47%), Gaps = 24/169 (14%)
Query 33 DYSGIGSSIGNIFQYDLMQSEKSQLQGARQLSDAKAMETLSNIDWGNLTAETRNYLRSTG 92
D+S + ++ + F+ L+ SE+S QG L A+ + L+ I G +AE N LRS
Sbjct 140 DFSLLSQAVDSFFKNKLL-SEQSTGQGLDNLLKARYGDELAQISIGKGSAEISN-LRS-- 195
Query 93 LARAQLGYVKEQQEVDNMAMTGLIMRAQRSGMLLDNEAKGILNKYLDQEQQLDLNVKAAD 152
Q N A T + L +A+ LNKYLD QQL L K A+
Sbjct 196 ------------QSARNYAETAVAS--------LTADAQRTLNKYLDMGQQLSLITKMAE 235
Query 153 YYQRMSAGYLSYaetkkaladealaaartrGQKISNEVASRIAESQIAA 201
Y + L+ A+ + +A+E A GQKISNE+A A+S I A
Sbjct 236 YSSITAGTELTKAKYRTEIANEIKTLAEANGQKISNEIARSTAQSLIDA 284
> Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.protein.BACEGG.02723
Length=418
Score = 47.4 bits (111), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 39/119 (33%), Positives = 69/119 (58%), Gaps = 2/119 (2%)
Query 98 LGYVKEQQEVDNMAMTGLIMRAQRSGMLLDNEAKG--ILNKYLDQEQQLDLNVKAADYYQ 155
LG ++ ++N++ I A G LL +A+G I+NK++ +QQ + +K A+ +
Sbjct 203 LGIDLRKKRLENLSTETNIKVALAQGALLGLQAEGQRIVNKFMPAQQQAEFFLKTANAFA 262
Query 156 RMSAGYLSYaetkkaladealaaartrGQKISNEVASRIAESQIAANIAANQSSEAYHN 214
+ AG LS A+ K + +AL A+ GQK++N +A R+A+ Q A A +++ AY+N
Sbjct 263 QYKAGKLSEAQVKTQIKQQALLEAQAAGQKLNNRLAERLADYQFKAMAAEYRANAAYYN 321
> Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.protein.BACEGG.02723
Length=418
Score = 47.4 bits (111), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 40/119 (34%), Positives = 70/119 (59%), Gaps = 2/119 (2%)
Query 98 LGYVKEQQEVDNMAMTGLIMRAQRSGMLLDNEAKG--ILNKYLDQEQQLDLNVKAADYYQ 155
LG ++ ++N++ I A G LL +A+G I+NK++ +QQ + +K A+ +
Sbjct 203 LGIDLRKKRLENLSTETNIKVALAQGALLGLQAEGQRIVNKFMPAQQQAEFFLKTANAFA 262
Query 156 RMSAGYLSYaetkkaladealaaartrGQKISNEVASRIAESQIAANIAANQSSEAYHN 214
+ AG LS A+ K + +AL A+T GQK++N +A R+A+ Q A A +++ AY+N
Sbjct 263 QYKAGKLSEAQVKTQIKQQALLEAQTVGQKLNNRLAERLADYQFKAMAAEYRANAAYYN 321
Lambda K H a alpha
0.313 0.128 0.356 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 23090192