bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-4_CDS_annotation_glimmer3.pl_2_3
Length=569
Score E
Sequences producing significant alignments: (Bits) Value
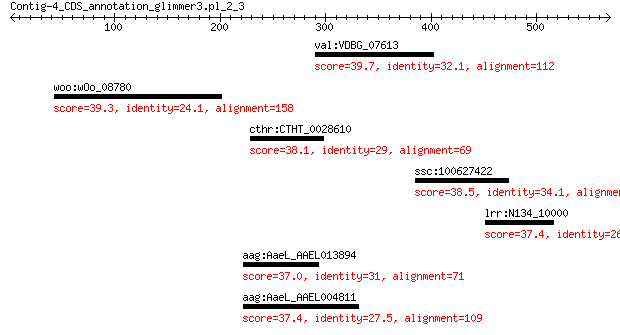
val:VDBG_07613 hypothetical protein 39.7 1.9
woo:wOo_08780 ATP-dependent exoDNase exonuclease V-subunit bet... 39.3 3.3
cthr:CTHT_0028610 6-phosphofructo-2-kinase/fructose-2,6-bispho... 38.1 4.9
ssc:100627422 ligand-dependent corepressor-like 38.5 5.2
lrr:N134_10000 diguanylate phosphodiesterase 37.4 7.1
aag:AaeL_AAEL013894 hypothetical protein 37.0 8.1
aag:AaeL_AAEL004811 hypothetical protein 37.4 8.7
> val:VDBG_07613 hypothetical protein
Length=431
Score = 39.7 bits (91), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 36/131 (27%), Positives = 60/131 (46%), Gaps = 20/131 (15%)
Query 290 NPSDSSTPNLSGSPLVLDVLALRRGEAL---------QRFREISLCTPANYRSQIKAHFG 340
P T ++ SP V + L R A Q ++I+ TPA +R++++ H G
Sbjct 216 KPGKQLTISVGASPQVASIENLTRPAAAGSPHGDHLRQTIQQITQNTPAGFRTKLELHAG 275
Query 341 VDVGSELSGMSTYIGGEASSLD-------ISEVV---NTNITESNEALIAGKGIGTGQFS 390
V ++ MST G ++D ++EVV N E+ EAL+A +G G+
Sbjct 276 VYSLLDVQQMSTQARGFLGAIDDEVAISVVAEVVSVYNNGERETPEALVAVGTLGLGREP 335
Query 391 DKFYAKDWGIL 401
Y+ WG++
Sbjct 336 CAAYS-GWGVI 345
> woo:wOo_08780 ATP-dependent exoDNase exonuclease V-subunit beta
RecB
Length=1126
Score = 39.3 bits (90), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 38/186 (20%), Positives = 72/186 (39%), Gaps = 33/186 (18%)
Query 43 EQHFTRTQPVNTSAYTRVREYY-------DWFWCPLHLLWRNAPEVIAQIQ--------- 86
+Q R Q N + +++Y+ DW C L +R+APE++ +
Sbjct 452 KQSIYRFQGANPHLFNYMQQYFHAKTGDRDWISCQLKKSFRSAPEILVFVDRIFNNFREE 511
Query 87 --------QNVQHASSYDGSVLLGSNMPCVSLSQLSKLLSSLKGKKNYFGFDR---SDLA 135
+++ H + G + + +P + L L KK++ DR +A
Sbjct 512 VSFINSEIKHIPHRKNDQGYIEIWPLLPRCKEEEQQALQIHLTDKKDHVISDRLLAQTIA 571
Query 136 YKILQYLRYGNVQTSSSTSGKNFGTSIPLSDRSYSQDYVFNHALSI-FPLLGYKKFCQDY 194
YKI +L + + + I + R+ DY+ N + P++G +DY
Sbjct 572 YKIHNWLSRRRILVAKDRHIEPRDIMILVRQRNVLVDYIINELRKVNVPVVG-----RDY 626
Query 195 FRFTQW 200
FR +
Sbjct 627 FRIMDY 632
> cthr:CTHT_0028610 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-like
protein
Length=534
Score = 38.1 bits (87), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 20/69 (29%), Positives = 37/69 (54%), Gaps = 3/69 (4%)
Query 229 SYLTHNTLIDMEYCNWNKDMFFGVLPDAQYGDASVVDISFGMSGQTVVASPSDISSRYTI 288
+YL H + +D+ + +D ++P A +A + I G+ +T+ +SP DI R +
Sbjct 422 AYLMHCSTMDIPKLKFPRDEIIEIIPAAYQNEAKRIHIP-GLDPKTIPSSPEDI--RIPV 478
Query 289 SNPSDSSTP 297
+PS +TP
Sbjct 479 PSPSGQATP 487
> ssc:100627422 ligand-dependent corepressor-like
Length=1714
Score = 38.5 bits (88), Expect = 5.2, Method: Composition-based stats.
Identities = 30/94 (32%), Positives = 44/94 (47%), Gaps = 9/94 (10%)
Query 385 GTGQFSDKFYAKDWGILMCI-YHSVPLLDYVLTSP---DPQLFLSENTSFPVPELDAIGL 440
G Q S K KD +C+ S P +D V+ S PQ + P PE +++
Sbjct 342 GCAQLSTKHKEKD---TVCLNMKSPPSVDLVIDSSGSHSPQHATEQALKEPPPETNSVDG 398
Query 441 ESIPLSCYSNSSLEIPITNPNVDAA--SLTMGYL 472
PL+ S E+P T PN +++ S T+GYL
Sbjct 399 RENPLTVVQKDSSELPTTKPNSESSVDSSTLGYL 432
> lrr:N134_10000 diguanylate phosphodiesterase
Length=265
Score = 37.4 bits (85), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 17/65 (26%), Positives = 33/65 (51%), Gaps = 1/65 (2%)
Query 451 SSLEIPITNPNVDAASLTMGYLPRYYAWKTSLDYVLGAFTTTEKEWVAPITASLWSKMLL 510
L+I +T NVD++ +T L RY + T L + + +F + W+ IT + W + +
Sbjct 166 KQLQIKVTIENVDSSKMTYNLLQRYLPYITYLKFNIHSFNKSANHWI-DITLAQWQRRIA 224
Query 511 PVTVD 515
++
Sbjct 225 IFNIE 229
> aag:AaeL_AAEL013894 hypothetical protein
Length=228
Score = 37.0 bits (84), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 22/71 (31%), Positives = 32/71 (45%), Gaps = 0/71 (0%)
Query 222 LPDTFSTSYLTHNTLIDMEYCNWNKDMFFGVLPDAQYGDASVVDISFGMSGQTVVASPSD 281
L D + SYLT + +E N + +GV A + VV + +SG V+ P D
Sbjct 52 LADVYLESYLTRTQVSRIERFNETALVHWGVCMPASCSERDVVQVVEAISGSIAVSIPKD 111
Query 282 ISSRYTISNPS 292
R I+ PS
Sbjct 112 ACHREVIAEPS 122
> aag:AaeL_AAEL004811 hypothetical protein
Length=536
Score = 37.4 bits (85), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 30/109 (28%), Positives = 46/109 (42%), Gaps = 5/109 (5%)
Query 222 LPDTFSTSYLTHNTLIDMEYCNWNKDMFFGVLPDAQYGDASVVDISFGMSGQTVVASPSD 281
L D + SYLT + +E N + +GV A + VV + +SG V+ P D
Sbjct 47 LADVYLESYLTRTQVSRIERFNETALVHWGVCMPASCSERDVVQVVEAISGSRAVSIPKD 106
Query 282 ISSRYTISNPSDSSTPNLSGSPLVLDVLALRRGEALQRFREISLCTPAN 330
R I+ PS +S + ++AL +Q S+CT N
Sbjct 107 ACHREVIAEPSTLDIVYVSIIMFFVLMVALSTLYHIQ-----SICTRRN 150
Lambda K H a alpha
0.318 0.134 0.409 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1296212451312