bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-4_CDS_annotation_glimmer3.pl_2_1
Length=128
Score E
Sequences producing significant alignments: (Bits) Value
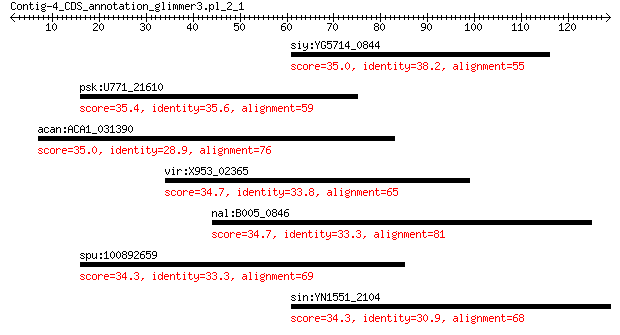
siy:YG5714_0844 hypothetical protein 35.0 0.88
psk:U771_21610 dsba oxidoreductase 35.4 1.5
acan:ACA1_031390 hypothetical protein 35.0 2.8
vir:X953_02365 hypothetical protein 34.7 3.5
nal:B005_0846 sucA; oxoglutarate dehydrogenase (succinyl-trans... 34.7 4.4
spu:100892659 uncharacterized LOC100892659 34.3 5.2
sin:YN1551_2104 homogentisate 12-dioxygenase 34.3 5.9
> siy:YG5714_0844 hypothetical protein
Length=93
Score = 35.0 bits (79), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 30/55 (55%), Gaps = 4/55 (7%)
Query 61 LKNQLSLQELVFAELGYSDELFDSFYVKPHIKVLKNAYIDKWKDVNYKEVHYFRV 115
+ +L L EL AE+ YS+E VK + K +K +DKW+ VN+ YFR
Sbjct 40 IAEELKLNEL-KAEITYSEEATTYVEVKLYKKAIKGTLLDKWRRVNF---SYFRC 90
> psk:U771_21610 dsba oxidoreductase
Length=214
Score = 35.4 bits (80), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 31/65 (48%), Gaps = 6/65 (9%)
Query 16 LNWDVPIGKISRFFYRFNRF------EAMKGSLRSKLKAVSLFYDYRDYQSLKNQLSLQE 69
LN D+P+ + R YR +F +AM + KAV L +DY + N L+
Sbjct 48 LNPDMPVQGMDRKVYRSGKFGSWARSQAMDAQVTHAGKAVGLEFDYERVKKTPNTLAAHR 107
Query 70 LVFAE 74
LV+ E
Sbjct 108 LVWRE 112
> acan:ACA1_031390 hypothetical protein
Length=296
Score = 35.0 bits (79), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 22/76 (29%), Positives = 32/76 (42%), Gaps = 0/76 (0%)
Query 7 IVRLDGYSFLNWDVPIGKISRFFYRFNRFEAMKGSLRSKLKAVSLFYDYRDYQSLKNQLS 66
I L G NWDV +GKI Y + +A+ GSL + SL N
Sbjct 134 IRALGGLGQKNWDVAVGKIGNRDYTLRQIQAVVGSLNDSRAWSCMVSACVSCPSLLNTAY 193
Query 67 LQELVFAELGYSDELF 82
L + + A++ S L+
Sbjct 194 LPQTLDAQMNLSAALY 209
> vir:X953_02365 hypothetical protein
Length=408
Score = 34.7 bits (78), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 22/67 (33%), Positives = 40/67 (60%), Gaps = 3/67 (4%)
Query 34 RFEAMKGSLRSKLKAVSLFYDYRDYQSLKN-QLSLQELVFA-ELGYSDELFDSFYVKPHI 91
+F+ KG ++ +K + L DYR S + ++ ++ V++ EL Y +L +SFY +P I
Sbjct 36 KFKVEKGDIKD-IKNLKLVGDYRQGMSAEGVAITSEDTVYSSELSYFSQLNNSFYTEPAI 94
Query 92 KVLKNAY 98
+ L+N Y
Sbjct 95 QQLQNNY 101
> nal:B005_0846 sucA; oxoglutarate dehydrogenase (succinyl-transferring),
E1 component (EC:1.2.4.2)
Length=1208
Score = 34.7 bits (78), Expect = 4.4, Method: Composition-based stats.
Identities = 27/92 (29%), Positives = 48/92 (52%), Gaps = 13/92 (14%)
Query 44 SKLKAVSLFYDYRDYQSLKNQLSLQ---ELVFAELGYSDELFDSFYVKPH--------IK 92
SK+ ++ YD+R Q ++ L+ +L+ E G+ DE+F+S + P+ IK
Sbjct 294 SKVMTLTSTYDHRIIQGAQSGEFLRRIHQLLLGEDGFYDEIFNSLRI-PYEPVRWVQDIK 352
Query 93 VLKNAYIDKWKDVNYKEVHYFRVKHKVLNDEN 124
V K +DK V + +H +RV+ ++ D N
Sbjct 353 VNKATQLDKTTRVQ-ELIHAYRVRGHLMADTN 383
> spu:100892659 uncharacterized LOC100892659
Length=1860
Score = 34.3 bits (77), Expect = 5.2, Method: Composition-based stats.
Identities = 23/72 (32%), Positives = 37/72 (51%), Gaps = 7/72 (10%)
Query 16 LNWDVPIGKISRFFYRFNRFEAMKGSLRSKLKAVSLFYDYRDYQS---LKNQLSLQELVF 72
+ WDV ++ F Y+F + + + R L +S YD Y S LK ++ LQEL
Sbjct 1061 ITWDV---EMDSFLYKFTPRDKPR-TRRGLLSVISSVYDPLGYASPFVLKAKMILQELTR 1116
Query 73 AELGYSDELFDS 84
LG+ D++ D+
Sbjct 1117 MNLGWDDKIPDA 1128
> sin:YN1551_2104 homogentisate 12-dioxygenase
Length=384
Score = 34.3 bits (77), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 21/73 (29%), Positives = 39/73 (53%), Gaps = 8/73 (11%)
Query 61 LKNQLSLQELVFAELGYSDELFDSFYVKPHIKVLK-----NAYIDKWKDVNYKEVHYFRV 115
+K+ L L+E VF G+S +++ P +++K N+ I++WKD YK + ++
Sbjct 19 MKDNLLLREEVFGLNGFSGRYSLLYHLNPPTRIVKVGEWRNSVIEEWKDAGYK---HHKL 75
Query 116 KHKVLNDENNIFL 128
L N++FL
Sbjct 76 STSKLRKSNDVFL 88
Lambda K H a alpha
0.326 0.142 0.430 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 128634530802