bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-35_CDS_annotation_glimmer3.pl_2_6
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
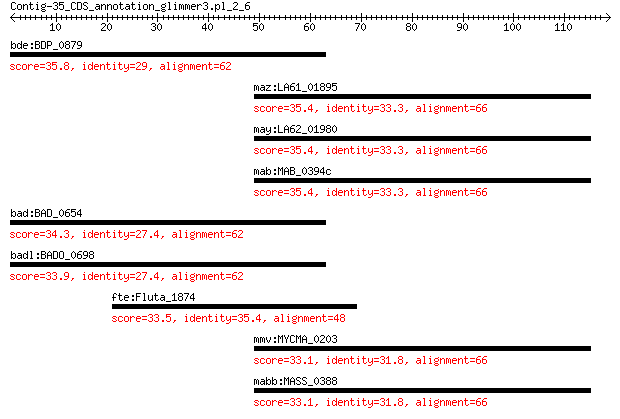
bde:BDP_0879 histidine kinase-like protein (EC:2.7.13.3) 35.8 1.2
maz:LA61_01895 hypothetical protein 35.4 1.5
may:LA62_01980 hypothetical protein 35.4 1.5
mab:MAB_0394c hypothetical protein 35.4 1.5
bad:BAD_0654 histidine kinase-like protein 34.3 4.7
badl:BADO_0698 histidine kinase-like protein 33.9 5.4
fte:Fluta_1874 Crp/Fnr family transcriptional regulator 33.5 5.7
mmv:MYCMA_0203 hypothetical protein 33.1 9.3
mabb:MASS_0388 hypothetical protein 33.1 9.6
> bde:BDP_0879 histidine kinase-like protein (EC:2.7.13.3)
Length=508
Score = 35.8 bits (81), Expect = 1.2, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 33/64 (52%), Gaps = 2/64 (3%)
Query 1 VQNKFLTYKNDNIMKYSFPTKNN--DRLKSVEIYEGESIETKCARILQNKEPITDTAPII 58
V + F+ D I++Y+ P + RL SV +GE + R+L +P+ +T P++
Sbjct 187 VSDGFIVLTVDGIVRYASPNAISCFRRLGSVSTMQGEYLSEIGTRLLHENDPVLETLPLV 246
Query 59 YTAK 62
+ K
Sbjct 247 LSGK 250
> maz:LA61_01895 hypothetical protein
Length=442
Score = 35.4 bits (80), Expect = 1.5, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 35/68 (51%), Gaps = 4/68 (6%)
Query 49 EPITDTAPIIYTAKEDGVLPAYNIRTDR--FDIAMDAYDKITRSSAKKEIAPKPEDFGNV 106
EP+ P + A +G L ++R R FD+ DA D TR +A+K++A +FG
Sbjct 100 EPLRPALPYVAHALREGRLGDEHVRIIRRFFDVLPDAVDADTREAAEKQLASMGTEFG-- 157
Query 107 PNKTEGGS 114
P + G+
Sbjct 158 PEQLRAGA 165
> may:LA62_01980 hypothetical protein
Length=442
Score = 35.4 bits (80), Expect = 1.5, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 35/68 (51%), Gaps = 4/68 (6%)
Query 49 EPITDTAPIIYTAKEDGVLPAYNIRTDR--FDIAMDAYDKITRSSAKKEIAPKPEDFGNV 106
EP+ P + A +G L ++R R FD+ DA D TR +A+K++A +FG
Sbjct 100 EPLRPALPYVAHALREGRLGDEHVRIIRRFFDVLPDAVDADTREAAEKQLASMGTEFG-- 157
Query 107 PNKTEGGS 114
P + G+
Sbjct 158 PEQLRAGA 165
> mab:MAB_0394c hypothetical protein
Length=442
Score = 35.4 bits (80), Expect = 1.5, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 35/68 (51%), Gaps = 4/68 (6%)
Query 49 EPITDTAPIIYTAKEDGVLPAYNIRTDR--FDIAMDAYDKITRSSAKKEIAPKPEDFGNV 106
EP+ P + A +G L ++R R FD+ DA D TR +A+K++A +FG
Sbjct 100 EPLRPALPYVAHALREGRLGDEHVRIIRRFFDVLPDAVDADTREAAEKQLASMGTEFG-- 157
Query 107 PNKTEGGS 114
P + G+
Sbjct 158 PEQLRAGA 165
> bad:BAD_0654 histidine kinase-like protein
Length=508
Score = 34.3 bits (77), Expect = 4.7, Method: Composition-based stats.
Identities = 17/64 (27%), Positives = 33/64 (52%), Gaps = 2/64 (3%)
Query 1 VQNKFLTYKNDNIMKYSFPTKNN--DRLKSVEIYEGESIETKCARILQNKEPITDTAPII 58
V + F+ D I++Y+ P + RL SV +GE + ++L +P+ +T P++
Sbjct 186 VADGFIVLTVDGIVRYASPNAISCFRRLGSVSTMQGEYLSEIGTKLLHENDPVLETLPLV 245
Query 59 YTAK 62
+ K
Sbjct 246 LSGK 249
> badl:BADO_0698 histidine kinase-like protein
Length=495
Score = 33.9 bits (76), Expect = 5.4, Method: Composition-based stats.
Identities = 17/64 (27%), Positives = 33/64 (52%), Gaps = 2/64 (3%)
Query 1 VQNKFLTYKNDNIMKYSFPTKNN--DRLKSVEIYEGESIETKCARILQNKEPITDTAPII 58
V + F+ D I++Y+ P + RL SV +GE + ++L +P+ +T P++
Sbjct 173 VADGFIVLTVDGIVRYASPNAISCFRRLGSVSTMQGEYLSEIGTKLLHENDPVLETLPLV 232
Query 59 YTAK 62
+ K
Sbjct 233 LSGK 236
> fte:Fluta_1874 Crp/Fnr family transcriptional regulator
Length=192
Score = 33.5 bits (75), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 29/48 (60%), Gaps = 3/48 (6%)
Query 21 KNNDRLKSVEIYEGESIETKCARILQNKEPITDTAPIIYTAKEDGVLP 68
K +RLK ++ ES+ET+ R+++N+E + AP+ + A GV P
Sbjct 133 KAVERLKE---FQTESLETRYLRLIENQEELFKAAPLQHIASYLGVKP 177
> mmv:MYCMA_0203 hypothetical protein
Length=442
Score = 33.1 bits (74), Expect = 9.3, Method: Composition-based stats.
Identities = 21/68 (31%), Positives = 34/68 (50%), Gaps = 4/68 (6%)
Query 49 EPITDTAPIIYTAKEDGVLPAYNIRTDR--FDIAMDAYDKITRSSAKKEIAPKPEDFGNV 106
EP+ P + A +G L ++R R FD+ DA D TR +A++ +A +FG
Sbjct 100 EPLRPALPYVAHALREGRLGDEHVRIIRRFFDVLPDAVDADTREAAEERLASMGTEFG-- 157
Query 107 PNKTEGGS 114
P + G+
Sbjct 158 PEQLRAGA 165
> mabb:MASS_0388 hypothetical protein
Length=442
Score = 33.1 bits (74), Expect = 9.6, Method: Composition-based stats.
Identities = 21/68 (31%), Positives = 34/68 (50%), Gaps = 4/68 (6%)
Query 49 EPITDTAPIIYTAKEDGVLPAYNIRTDR--FDIAMDAYDKITRSSAKKEIAPKPEDFGNV 106
EP+ P + A +G L ++R R FD+ DA D TR +A++ +A +FG
Sbjct 100 EPLRPALPYVAHALREGRLGDEHVRIIRRFFDVLPDAVDADTREAAEERLASMGTEFG-- 157
Query 107 PNKTEGGS 114
P + G+
Sbjct 158 PEQLRAGA 165
Lambda K H a alpha
0.310 0.130 0.366 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 129080580354